

You can:
| Name | Beta-1 adrenergic receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | ADRB1 |
| Synonym | Adrenergic receptor beta 1 Adrb-1 ADRB1R adrenergic receptor beta 1-AR [ Show all ] |
| Disease | Heart failure; Cardiogenic shock Heart failure Glaucoma Diabetes Coronary artery disease [ Show all ] |
| Length | 477 |
| Amino acid sequence | MGAGVLVLGASEPGNLSSAAPLPDGAATAARLLVPASPPASLLPPASESPEPLSQQWTAGMGLLMALIVLLIVAGNVLVIVAIAKTPRLQTLTNLFIMSLASADLVMGLLVVPFGATIVVWGRWEYGSFFCELWTSVDVLCVTASIETLCVIALDRYLAITSPFRYQSLLTRARARGLVCTVWAISALVSFLPILMHWWRAESDEARRCYNDPKCCDFVTNRAYAIASSVVSFYVPLCIMAFVYLRVFREAQKQVKKIDSCERRFLGGPARPPSPSPSPVPAPAPPPGPPRPAAAAATAPLANGRAGKRRPSRLVALREQKALKTLGIIMGVFTLCWLPFFLANVVKAFHRELVPDRLFVFFNWLGYANSAFNPIIYCRSPDFRKAFQRLLCCARRAARRRHATHGDRPRASGCLARPGPPPSPGAASDDDDDDVVGATPPARLLEPWAGCNGGAAADSDSSLDEPCRPGFASESKV |
| UniProt | P08588 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P08588 |
| 3D structure model | This predicted structure model is from GPCR-EXP P08588. |
| BioLiP | N/A |
| Therapeutic Target Database | T44068 |
| ChEMBL | CHEMBL213 |
| IUPHAR | 28 |
| DrugBank | BE0000172 |
| Name | propranolol |
|---|---|
| Molecular formula | C16H21NO2 |
| IUPAC name | 1-naphthalen-1-yloxy-3-(propan-2-ylamino)propan-2-ol |
| Molecular weight | 259.349 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 2 |
| XlogP | 3.0 |
| Synonyms | (S)-(-)-PROPRANOLOL Spectrum2_001301 1-(alpha-naphthoxy)-3-(isopropylamino)-2-propanol STK735510 DB00571 [ Show all ] |
| Inchi Key | AQHHHDLHHXJYJD-UHFFFAOYSA-N |
| Inchi ID | InChI=1S/C16H21NO2/c1-12(2)17-10-14(18)11-19-16-9-5-7-13-6-3-4-8-15(13)16/h3-9,12,14,17-18H,10-11H2,1-2H3 |
| PubChem CID | 4946 |
| ChEMBL | CHEMBL27 |
| IUPHAR | 564 |
| BindingDB | 25761 |
| DrugBank | DB00571 |
Structure | 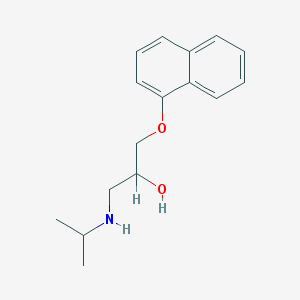 |
| Lipinski's druglikeness | This ligand satisfies Lipinski's rule of five. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| N/A | N/A | DrugBank | |
| Contractile force | 7.6 - | PMID2861287 | ChEMBL |
| DR10 | 0.13 mg kg-1 | PMID6124636 | ChEMBL |
| IC50 | 2.543 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| IC50 | 18.0 nM | PMID1976812 | BindingDB,ChEMBL |
| IC50 | 251.19 nM | PMID26565745 | ChEMBL |
| ID50 | 0.069 mg.kg-1 | PMID3001305 | ChEMBL |
| ID50 | 0.08 umol.kg-1 | PMID2892934 | ChEMBL |
| Kd | 1.8 nM | PMID21870877 | BindingDB |
| Kd | 1.82 nM | PMID21870877 | ChEMBL |
| Kd | 6.0 nM | PMID21870877 | BindingDB |
| Kd | 6.026 nM | PMID21870877 | ChEMBL |
| Kd | 6.9 nM | PMID15655528 | BindingDB |
| Kd | 199.53 nM | PMID21870877 | ChEMBL |
| Kd | 200.0 nM | PMID21870877 | BindingDB |
| Ki | 0.02 nM | PMID8935801 | PDSP,BindingDB |
| Ki | 1.468 nM | DrugMatrix in vitro pharmacology data | ChEMBL |
| Ki | 1.8 nM | PMID14730417 | PDSP,BindingDB |
| Ki | 2.3 nM | PMID8093626 | BindingDB,ChEMBL |
| Ki | 2.69 nM | PMID7915318 | PDSP,BindingDB |
zhanglab![]() zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
zhanggroup.org | (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218