

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MENSTEVTEFILLGLTDDPNLQIPLLLAFLFIYLITLLGNGGMMVIIHSDSHLHTPMYFFLSNLSLVDLGYSSAVAPKTVAALRSGDKAISYDGCAAQFFFFVGFATVECYLLASMAYDRHAAVCRPLHYTTTMTAGVCALLATGSYVSGFLNASIHAAGTFRLSFCGSNEINHFFCDIPPLLALSCSDTRISKLVVFVAGFNVFFTLLVILISYFFICITIQRMHSAEGQKKVFSTCASHLTALSIFYGTIIFMYLQPNSSQSVDTDKIASVFYTVVIPMLNPLIYSLRNKEVKSALWKILNKLYPQY | |
| CCCCCCCHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHHCCCHHHHHHHHHHHHHHCCCC | |
| 998962110268648898252899999999999999998879999985188877749899877998844721247099999870489788589999999999999999999999998651452062202880267889999999999999999999999985540899991385416808888771568518789999999999999999999999999998026775766443216588899799997432145837899988878988998822003124357450465989999999998645579 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MENSTEVTEFILLGLTDDPNLQIPLLLAFLFIYLITLLGNGGMMVIIHSDSHLHTPMYFFLSNLSLVDLGYSSAVAPKTVAALRSGDKAISYDGCAAQFFFFVGFATVECYLLASMAYDRHAAVCRPLHYTTTMTAGVCALLATGSYVSGFLNASIHAAGTFRLSFCGSNEINHFFCDIPPLLALSCSDTRISKLVVFVAGFNVFFTLLVILISYFFICITIQRMHSAEGQKKVFSTCASHLTALSIFYGTIIFMYLQPNSSQSVDTDKIASVFYTVVIPMLNPLIYSLRNKEVKSALWKILNKLYPQY | |
| 862403011000000043240101001303331331333332002001000300000000020000000100000003000100144320103000100100000000001000000010000002001000300330000001101010010010100000202014312010000032100100013231101000101323331331033033100100010302632310100000021001023201200000032431244410000110320023003000020330140033005423657 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCHHSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCCHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHHCCCHHHHHHHHHHHHHHCCCC MENSTEVTEFILLGLTDDPNLQIPLLLAFLFIYLITLLGNGGMMVIIHSDSHLHTPMYFFLSNLSLVDLGYSSAVAPKTVAALRSGDKAISYDGCAAQFFFFVGFATVECYLLASMAYDRHAAVCRPLHYTTTMTAGVCALLATGSYVSGFLNASIHAAGTFRLSFCGSNEINHFFCDIPPLLALSCSDTRISKLVVFVAGFNVFFTLLVILISYFFICITIQRMHSAEGQKKVFSTCASHLTALSIFYGTIIFMYLQPNSSQSVDTDKIASVFYTVVIPMLNPLIYSLRNKEVKSALWKILNKLYPQY | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.21 | 0.90 | 3.63 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.24 | 0.88 | 2.01 | Download | -ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM-------- | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.22 | 0.86 | 2.01 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-------PPFFCVVNT----------------DHILYTVYSTVGAFY---FPTLLLIALYGRIYVEARSRIIQKYAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK---- | |||||||||||||||||||
| 4 | 3uon | 0.19 | 0.19 | 0.89 | 1.55 | Download | ------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVG--VRTVEDGECYIQFF---------SNAAVTGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------ | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.23 | 0.91 | 1.25 | Download | TTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.21 | 0.90 | 3.78 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.23 | 0.85 | 1.71 | Download | -------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASECVVNT-----------------DHILYTVY--STVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--LAI-FDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF----- | |||||||||||||||||||
| 8 | 2z73A | 0.18 | 0.22 | 0.94 | 2.92 | Download | WYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGCSFDYISR----------------DSTTRSNILCMFILGF--FGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCC | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.21 | 0.90 | 5.25 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDV----------VPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.22 | 0.93 | 5.90 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFCVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
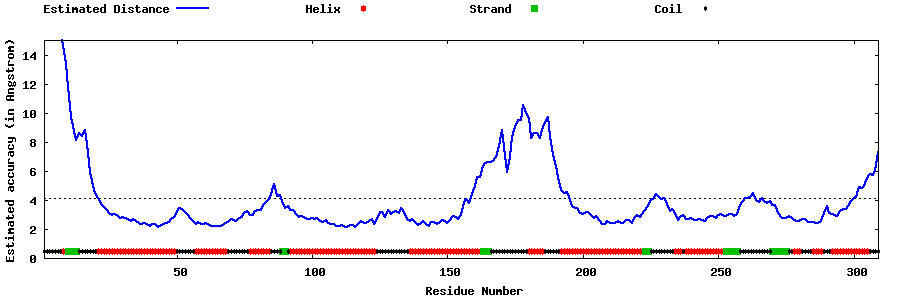 | |||
|
|
|||
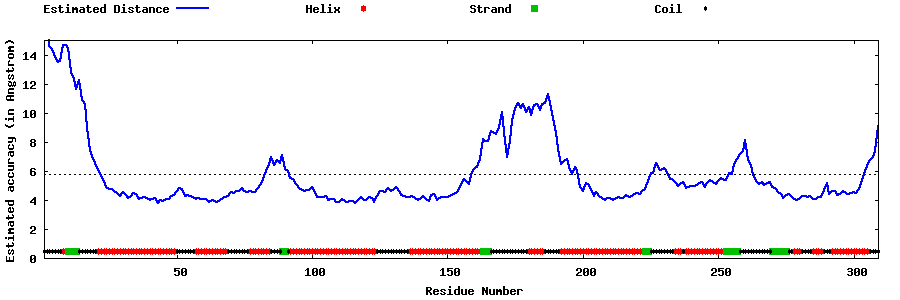 | |||
|
| |||
 | |||
|
| |||
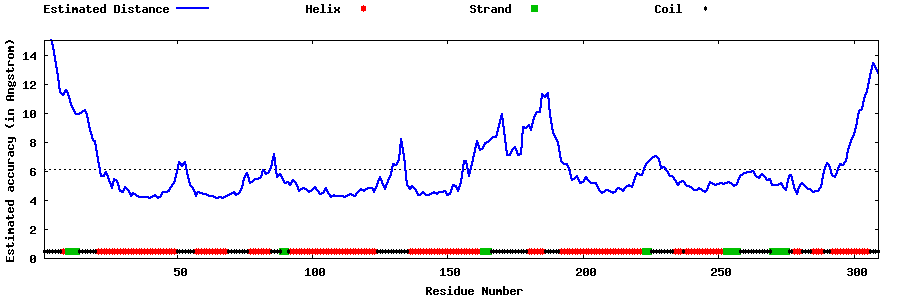 | |||
|
| |||
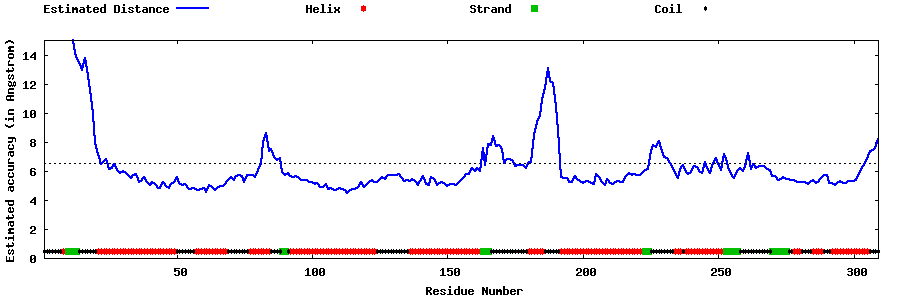 | |||
|
| |||