

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MKNRTSVTDFILLGLTDNPQLQVVIFSFLFLTYVLSVTGNLTIISLTLLDSHLKTPMYFFLRNFSLEISFTSVCNPRFLISILTGDKSISYNACAAQLFFFIFLGSTEFFLLASMSYDCYVAICKPLHYTTIMSDRICYQLIISSWLAGFLVIFPPLAMGLQLDFCDSNVIDHFTCDSAPLLQISCTDTSTLELMSFILALFTLISTLILVILSYTYIIRTILRIPSAQQRKKAFSTCSSHVIVVSISYGSCIFMYVKTSAKEGVALTKGVAILNTSVAPMLNPFIYTLRNQQVKQAFKDVLRKISHKKKKH | |
| CCCCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCHHHHHHHHHCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHCC | |
| 999851020357458998344999999999999999998999999997577767738888788988767741578999898733996870899999999999999999999999875511520655327821677699999999999999999999999816067899955883158488887714575199999999999999998999999999999999218652247631124385799689997414216868899877878868976243167502465321564989999999999876464159 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MKNRTSVTDFILLGLTDNPQLQVVIFSFLFLTYVLSVTGNLTIISLTLLDSHLKTPMYFFLRNFSLEISFTSVCNPRFLISILTGDKSISYNACAAQLFFFIFLGSTEFFLLASMSYDCYVAICKPLHYTTIMSDRICYQLIISSWLAGFLVIFPPLAMGLQLDFCDSNVIDHFTCDSAPLLQISCTDTSTLELMSFILALFTLISTLILVILSYTYIIRTILRIPSAQQRKKAFSTCSSHVIVVSISYGSCIFMYVKTSAKEGVALTKGVAILNTSVAPMLNPFIYTLRNQQVKQAFKDVLRKISHKKKKH | |
| 872403011000000033340001001303331332333231002002002300000000020001001010010032000001442200010000011000000110010000000000000021110102003300100002013201310302002003040033110100000210001000131312012003313313331331033033201200010313632310100111031002023201200001031633143410000200312033003000020430140033004202324668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCHHHHHHHHHCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHHHHHHHCC MKNRTSVTDFILLGLTDNPQLQVVIFSFLFLTYVLSVTGNLTIISLTLLDSHLKTPMYFFLRNFSLEISFTSVCNPRFLISILTGDKSISYNACAAQLFFFIFLGSTEFFLLASMSYDCYVAICKPLHYTTIMSDRICYQLIISSWLAGFLVIFPPLAMGLQLDFCDSNVIDHFTCDSAPLLQISCTDTSTLELMSFILALFTLISTLILVILSYTYIIRTILRIPSAQQRKKAFSTCSSHVIVVSISYGSCIFMYVKTSAKEGVALTKGVAILNTSVAPMLNPFIYTLRNQQVKQAFKDVLRKISHKKKKH | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.22 | 0.88 | 3.40 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADI-GVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLA-IIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL--RQ- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.22 | 0.89 | 2.22 | Download | -ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVDLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL--------GWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM----------- | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.22 | 0.88 | 1.97 | Download | --------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSSDIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAP-DQARMDIELAKTLVLILVVLIICWGPLLAIMKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 4 | 3uon | 0.16 | 0.17 | 0.88 | 1.55 | Download | ------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLAADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVG--VRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIAREKKVTRTILAILLAFIITWAPYNVMVLINTFPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM--------- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.21 | 0.90 | 1.24 | Download | TTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALDLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVF-F------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGADIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.22 | 0.88 | 3.65 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.22 | 0.83 | 1.71 | Download | ------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF-------- | |||||||||||||||||||
| 8 | 2z73A | 0.17 | 0.21 | 0.95 | 2.81 | Download | WYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCNDYI----------------SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFD | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.22 | 0.89 | 4.61 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 10 | 2ydoA | 0.15 | 0.21 | 0.93 | 5.50 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEP | |||||||||||||||||||
| ||||||||||||||||||||||||||
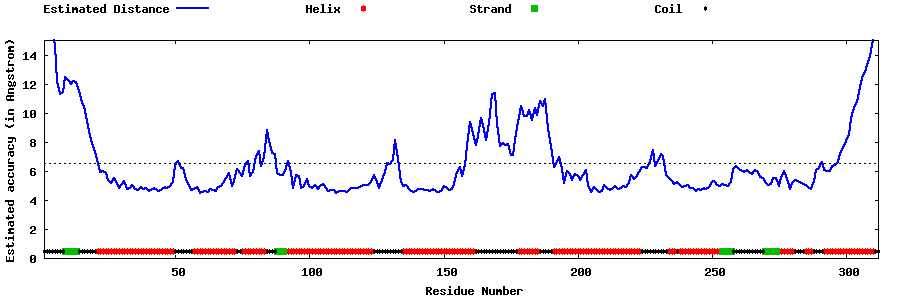
| Generated 3D models | Estimated local accuracy of models | ||
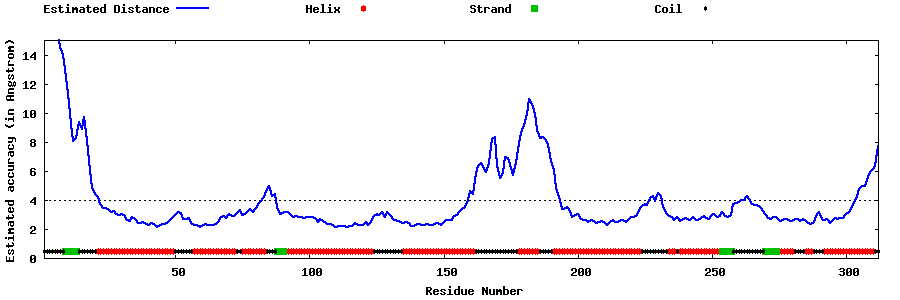 | |||
|
|
|||
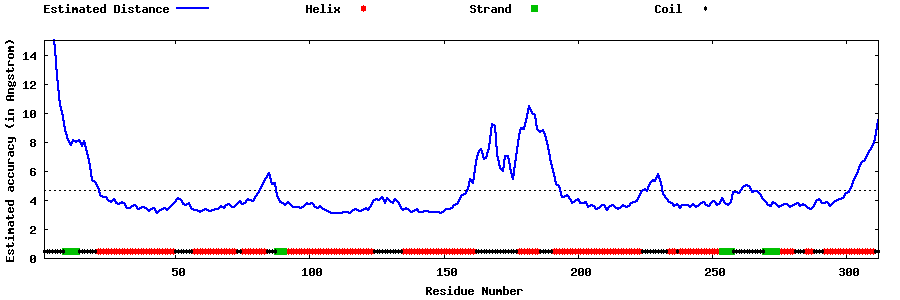 | |||
|
| |||
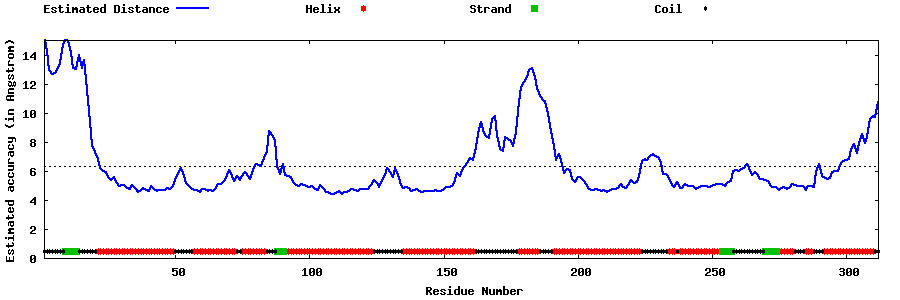 | |||
|
| |||
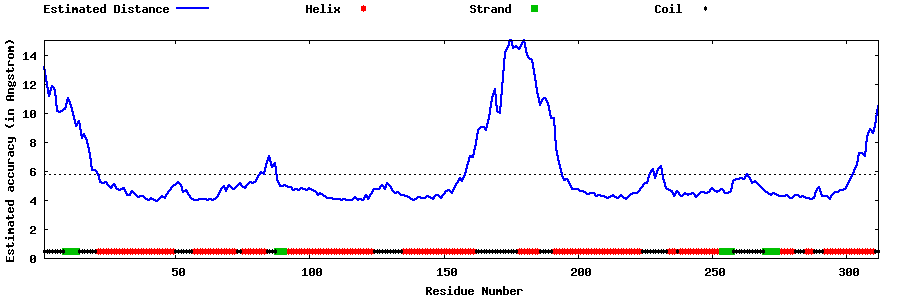 | |||
|
| |||
 | |||
|
| |||