

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MALPITNGTLFMPFVLTFIGIPGFESVQCWIGIPFCATYVIALIGNSLLLIIIKSEPSLHEPMYIFLATLGATDISLSTSIVPKMLDIFWFHLPEIYFDACLFQMWLIHTFQGIESGVLLAMALDRCVAICYPLRRAIVFTRQLVTYIVVGVTLRPAILVIPCLLLIKCHLKLYRTKLIYHTYCERVALVKLATEDVYINKVYGILGAFIVGGLDFIFITLSYIQIFITVFHLPLKEARLKVFNTCIPHIYVFFQFYLLAFFFIFYSQIWILYPIICTYHLVQSLPTGPTIPQPLYLWVKDQTH | |
| CCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHCCCCCCCCSSSSSCCCCC | |
| 9999989886831899980898976688999999999999999999998887737887542189999999999898998559999999956998688878999999999999999999999982305402566545103888899999999999999999889999973788899995277221346567883468289899999999999999999999999999999837888758888754149999999999998999976874468788489999988884278987877988467889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MALPITNGTLFMPFVLTFIGIPGFESVQCWIGIPFCATYVIALIGNSLLLIIIKSEPSLHEPMYIFLATLGATDISLSTSIVPKMLDIFWFHLPEIYFDACLFQMWLIHTFQGIESGVLLAMALDRCVAICYPLRRAIVFTRQLVTYIVVGVTLRPAILVIPCLLLIKCHLKLYRTKLIYHTYCERVALVKLATEDVYINKVYGILGAFIVGGLDFIFITLSYIQIFITVFHLPLKEARLKVFNTCIPHIYVFFQFYLLAFFFIFYSQIWILYPIICTYHLVQSLPTGPTIPQPLYLWVKDQTH | |
| 7603431314230310200000121320200021022012203321210000020141001000200010011000002002020000000204403040000001201310230110012003000000021010000003310010000003212012200100022104204320000000102100100023030011100200331333323103301220020003002460132012001001000100333133101001003300210001002210030033002000021358 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHCCCCCCCCSSSSSCCCCC MALPITNGTLFMPFVLTFIGIPGFESVQCWIGIPFCATYVIALIGNSLLLIIIKSEPSLHEPMYIFLATLGATDISLSTSIVPKMLDIFWFHLPEIYFDACLFQMWLIHTFQGIESGVLLAMALDRCVAICYPLRRAIVFTRQLVTYIVVGVTLRPAILVIPCLLLIKCHLKLYRTKLIYHTYCERVALVKLATEDVYINKVYGILGAFIVGGLDFIFITLSYIQIFITVFHLPLKEARLKVFNTCIPHIYVFFQFYLLAFFFIFYSQIWILYPIICTYHLVQSLPTGPTIPQPLYLWVKDQTH | |||||||||||||||||||||||||
| 1 | 5tgzA | 0.17 | 0.22 | 0.91 | 2.30 | Download | -GGRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDH | |||||||||||||||||||
| 2 | 5tgzA | 0.17 | 0.22 | 0.85 | 1.92 | Download | -------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWEKLQSVCSDIFPHID-------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLLLNSTVNPIIYALRSKD- | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.22 | 0.91 | 1.83 | Download | -------GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLNCEKLQSVC-SDIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDL | |||||||||||||||||||
| 4 | 3uon | 0.15 | 0.19 | 0.88 | 1.56 | Download | ------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFW-QFIVG--VRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPCYALCNATFK | |||||||||||||||||||
| 5 | 3uonA | 0.16 | 0.20 | 0.87 | 1.16 | Download | ------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQ---FIVGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPIPNTV-WTIGYWLCYINSTINPACYALCNATF | |||||||||||||||||||
| 6 | 5tgzA | 0.16 | 0.22 | 0.92 | 2.50 | Download | -GGRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSV-----------------CSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDL | |||||||||||||||||||
| 7 | 3d4s | 0.16 | 0.17 | 0.64 | 1.71 | Download | ----------------------------VGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRAT-----HQEAINCYAE----ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAK------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.21 | 0.84 | 1.86 | Download | ------NSDL---------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPI-------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYIS---------------------------DE | |||||||||||||||||||
| 9 | 5tgzA | 0.17 | 0.22 | 0.91 | 3.03 | Download | -GGRGENFMDIEC--FMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDL | |||||||||||||||||||
| 10 | 4gpoA | 0.15 | 0.21 | 0.89 | 4.66 | Download | -----------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMM-HWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDF | |||||||||||||||||||
| ||||||||||||||||||||||||||
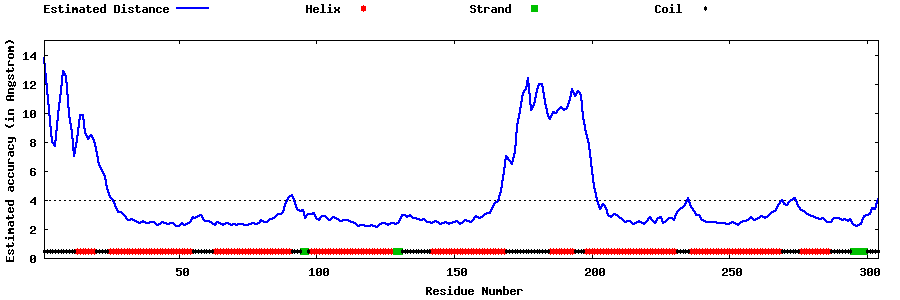
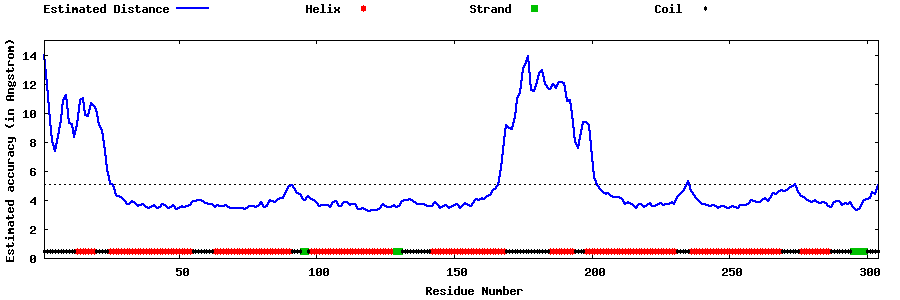
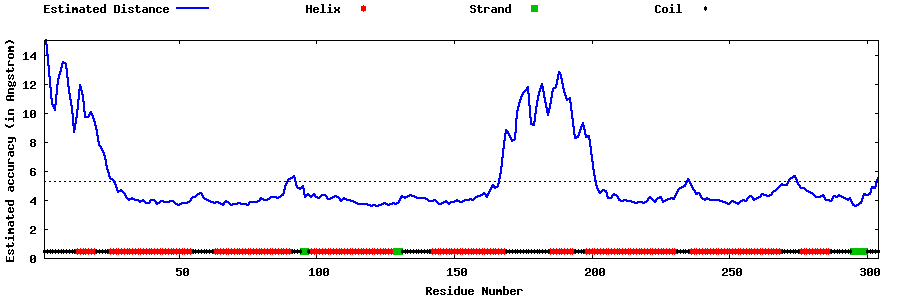
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||