

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MMIKKNASSEDFFILLGFSNWPQLEVVLFVVILIFYLMTLTGNLFIIILSYVDSHLHTPMYFFLSNLSFLDLCHTTSSIPQLLVNLRGPEKTISYAGCMVQLYFVLALGIAECVLLVVMSYDRYVAVCRPLHYTVLMHPRFCHLLAAASWVIGFTISALHSSFTFWVPLCGHRLVDHFFCEVPALLRLSCVDTHANELTLMVMSSIFVLIPLILILTAYGAIARAVLSMQSTTGLQKVFRTCGAHLMVVSLFFIPVMCMYLQPPSENSPDQGKFIALFYTVVTPSLNPLIYTLRNKHVKGAAKRLLGWEWGK | |
| CCCCCCCSSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCSSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHHCCC | |
| 998888701146689669998417999999999999999998899999997488756758999878999877776257899989846899668389999999999999999999999998650776165444771358889999999999999999999999981438999894688604838999886146529999999999999999899999999999999822856054661310648689979998743550681789999887881899744061112125643256299999999997553289 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MMIKKNASSEDFFILLGFSNWPQLEVVLFVVILIFYLMTLTGNLFIIILSYVDSHLHTPMYFFLSNLSFLDLCHTTSSIPQLLVNLRGPEKTISYAGCMVQLYFVLALGIAECVLLVVMSYDRYVAVCRPLHYTVLMHPRFCHLLAAASWVIGFTISALHSSFTFWVPLCGHRLVDHFFCEVPALLRLSCVDTHANELTLMVMSSIFVLIPLILILTAYGAIARAVLSMQSTTGLQKVFRTCGAHLMVVSLFFIPVMCMYLQPPSENSPDQGKFIALFYTVVTPSLNPLIYTLRNKHVKGAAKRLLGWEWGK | |
| 725562302011000000033240011001302331331333331002001001300000011020000100101001002000002165320100002201220130033002000100200000002102000200330000001100210031030001000001002614010000032100200021033001102331333133122000201220020001031351231011000000100101120010000103164254330000021033213300300101031013002200334368 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHCCHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCSSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHHHCCC MMIKKNASSEDFFILLGFSNWPQLEVVLFVVILIFYLMTLTGNLFIIILSYVDSHLHTPMYFFLSNLSFLDLCHTTSSIPQLLVNLRGPEKTISYAGCMVQLYFVLALGIAECVLLVVMSYDRYVAVCRPLHYTVLMHPRFCHLLAAASWVIGFTISALHSSFTFWVPLCGHRLVDHFFCEVPALLRLSCVDTHANELTLMVMSSIFVLIPLILILTAYGAIARAVLSMQSTTGLQKVFRTCGAHLMVVSLFFIPVMCMYLQPPSENSPDQGKFIALFYTVVTPSLNPLIYTLRNKHVKGAAKRLLGWEWGK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.21 | 0.20 | 0.89 | 3.62 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.22 | 0.88 | 2.22 | Download | --RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM------- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.22 | 0.88 | 2.20 | Download | GGRGENFMDIE-------CFMPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------ | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.24 | 0.90 | 1.55 | Download | --------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4yay | 0.19 | 0.26 | 0.91 | 1.24 | Download | LKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------ | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.20 | 0.88 | 3.84 | Download | ----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 7 | 4iaq | 0.22 | 0.26 | 0.84 | 1.72 | Download | ----------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 8 | 2z73A | 0.18 | 0.20 | 0.94 | 2.92 | Download | ----ENPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCNDYI----------------SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTC | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.20 | 0.89 | 4.82 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.23 | 0.92 | 5.69 | Download | ------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| ||||||||||||||||||||||||||
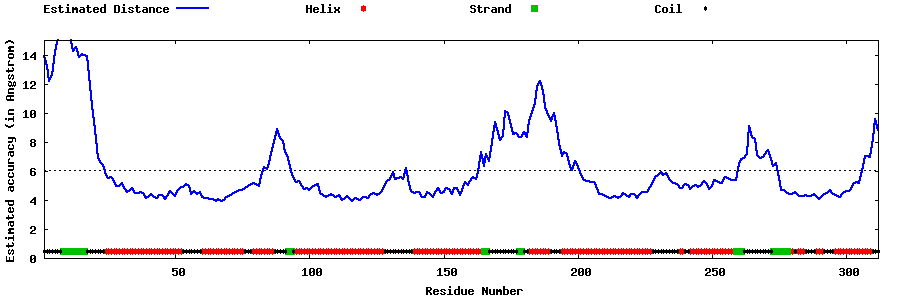
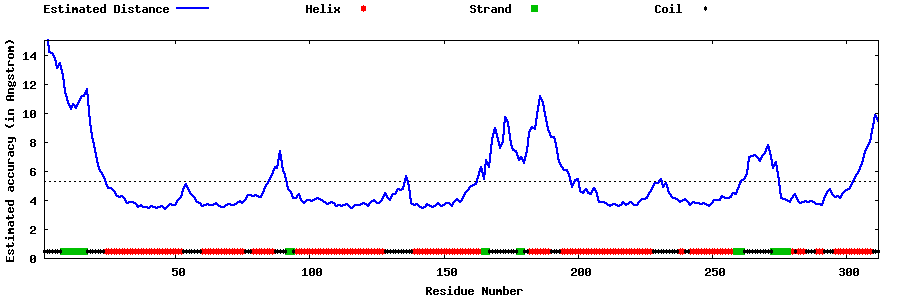
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||