

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MLLCFRFGNQSMKRENFTLITDFVFQGFSSFHEQQITLFGVFLALYILTLAGNIIIVTIIRMDLHLHTPMYFFLSMLSTSETVYTLVILPRMLSSLVGMSQPISLAGCATQMFFFVTFGITNCFLLTAMGYDRYVAICNPLRYMVIMNKRLRIQLVLGACSIGLIVAITQVTSVFRLPFCARKVPHFFCDIRPVMKLSCIDTTVNEILTLIISVLVLVVPMGLVFISYVLIISTILKIASVEGRKKAFATCASHLTVVIVHYSCASIAYLKPKSENTREHDQLISVTYTVITPLLNPVVYTLRNKEVKDALCRAVGGKFS | |
| CSSSCCCCCCCCCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCC | |
| 95553456133667778144576566699885179999999999999999989999898871887557489998889999788883187989898616997487899999999999999999999999986502511655447831578799999999999999999999999827127899998550682888888345750999999999999999959999999999999981387757676232165878997999861030478278999988788199986425232224653304669999999999734079 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MLLCFRFGNQSMKRENFTLITDFVFQGFSSFHEQQITLFGVFLALYILTLAGNIIIVTIIRMDLHLHTPMYFFLSMLSTSETVYTLVILPRMLSSLVGMSQPISLAGCATQMFFFVTFGITNCFLLTAMGYDRYVAICNPLRYMVIMNKRLRIQLVLGACSIGLIVAITQVTSVFRLPFCARKVPHFFCDIRPVMKLSCIDTTVNEILTLIISVLVLVVPMGLVFISYVLIISTILKIASVEGRKKAFATCASHLTVVIVHYSCASIAYLKPKSENTREHDQLISVTYTVITPLLNPVVYTLRNKEVKDALCRAVGGKFS | |
| 30102323444144402020000000000433501010013023312311332320010010033000000110200012032112033030001011643201020002011000000210010001001000000021020002003300000011002100310310010001020125501000002200010002203110211122123323313310330232000000103136323100000000100010112001000010216334534100003103320331030000204201300220044428 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CSSSCCCCCCCCCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCC MLLCFRFGNQSMKRENFTLITDFVFQGFSSFHEQQITLFGVFLALYILTLAGNIIIVTIIRMDLHLHTPMYFFLSMLSTSETVYTLVILPRMLSSLVGMSQPISLAGCATQMFFFVTFGITNCFLLTAMGYDRYVAICNPLRYMVIMNKRLRIQLVLGACSIGLIVAITQVTSVFRLPFCARKVPHFFCDIRPVMKLSCIDTTVNEILTLIISVLVLVVPMGLVFISYVLIISTILKIASVEGRKKAFATCASHLTVVIVHYSCASIAYLKPKSENTREHDQLISVTYTVITPLLNPVVYTLRNKEVKDALCRAVGGKFS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.16 | 0.17 | 0.86 | 3.41 | Download | -------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.22 | 0.87 | 2.11 | Download | ------------RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLCSDIFPHI------------------DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------ | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.22 | 0.86 | 2.22 | Download | ---------------------GGRGENFMDIPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCQSVCSDIFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAP-DQARMDIELAKTLVLILVVLIICWGPLLADVFGKMNKLIKTVFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.21 | 0.88 | 1.54 | Download | ------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR--EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 5 | 4yay | 0.15 | 0.20 | 0.92 | 1.25 | Download | KVKEAQAAAEQLKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF-----IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----- | |||||||||||||||||||
| 6 | 3emlA | 0.16 | 0.17 | 0.86 | 3.66 | Download | --------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.16 | 0.19 | 0.81 | 1.72 | Download | --------------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASE---------CVV----------NTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLG---IILGAFIVCWLIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 8 | 2z73A | 0.16 | 0.21 | 0.92 | 2.64 | Download | --------------ENPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLEGVLCNDY-----------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLT | |||||||||||||||||||
| 9 | 3emlA | 0.14 | 0.17 | 0.87 | 4.87 | Download | -------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLG-------WNNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 10 | 2ydoA | 0.14 | 0.17 | 0.89 | 5.53 | Download | ----------------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| ||||||||||||||||||||||||||
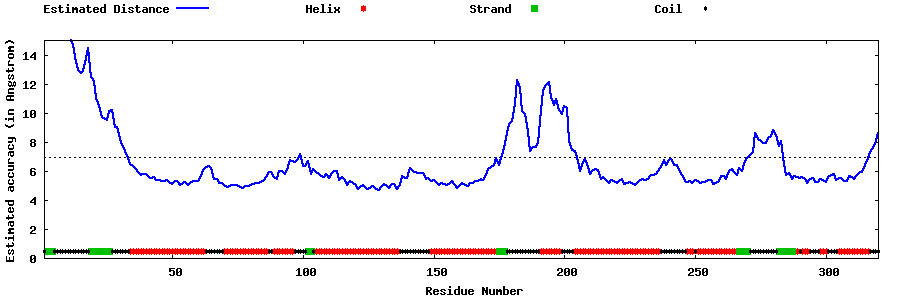
| Generated 3D models | Estimated local accuracy of models | ||
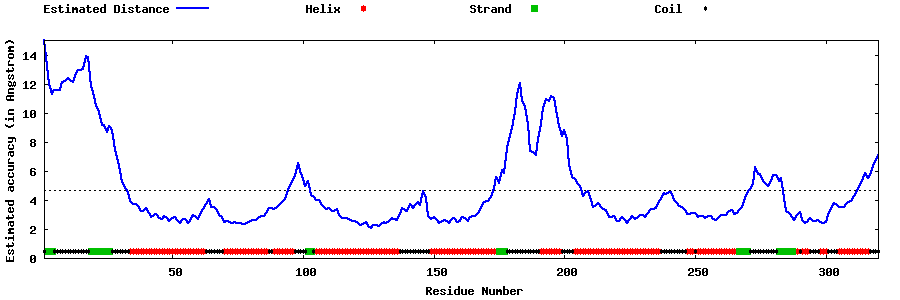 | |||
|
|
|||
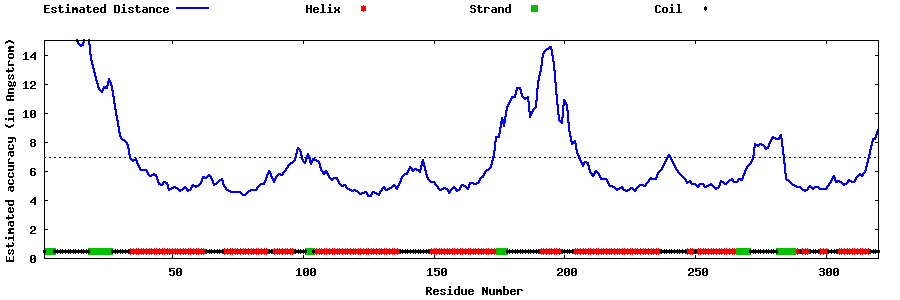 | |||
|
| |||
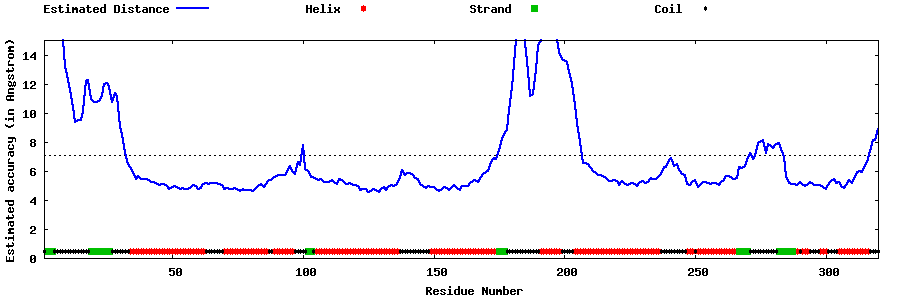 | |||
|
| |||
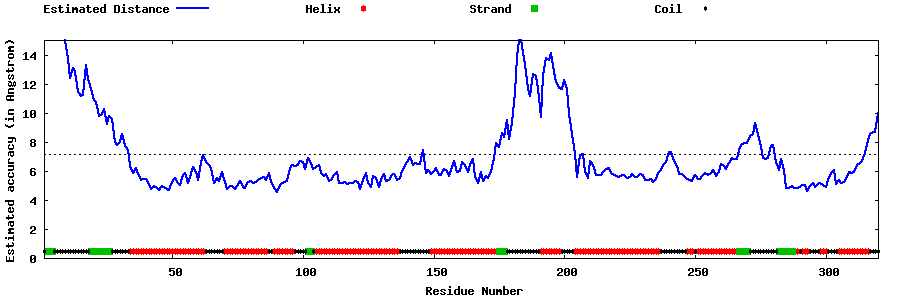 | |||
|
| |||
 | |||
|
| |||