

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 | |
| | | | | | | | | | | | | | | | | | | | | | | | | |
| MMDVNSSGRPDLYGHLRSFLLPEVGRGLPDLSPDGGADPVAGSWAPHLLSEVTASPAPTWDAPPDNASGCGEQINYGRVEKVVIGSILTLITLLTIAGNCLVVISVCFVKKLRQPSNYLIVSLALADLSVAVAVMPFVSVTDLIGGKWIFGHFFCNVFIAMDVMCCTASIMTLCVISIDRYLGITRPLTYPVRQNGKCMAKMILSVWLLSASITLPPLFGWAQNVNDDKVCLISQDFGYTIYSTAVAFYIPMSVMLFMYYQIYKAARKSAAKHKFPGFPRVEPDSVIALNGIVKLQKEVEECANLSRLLKHERKNISIFKREQKAATTLGIIVGAFTVCWLPFFLLSTARPFICGTSCSCIPLWVERTFLWLGYANSLINPFIYAFFNRDLRTTYRSLLQCQYRNINRKLSAAGMHEALKLAERPERPEFVLRACTRRVLLRPEKRPPVSVWVLQSPDHHNWLADKMLTTVEKKVMIHD | |
| CCCCCCCCCCCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCSSSSCC | |
| 97778787832111566766799999889999998788777777664223467788877778876556767788888899999999999999999996887278777557887567899999999999999998439999999649765777899999999999999999999999986466066410488656489899979999999999999999738864789983798547447778999999999999999999999999988776421322233233211134554444444322233343221013435553012012446689999999987999999999982764235561799999999999998160596860889999999998386467888887656554331245578899760344788644567889998766788887657554642435564576059 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 | |
| | | | | | | | | | | | | | | | | | | | | | | | | |
| MMDVNSSGRPDLYGHLRSFLLPEVGRGLPDLSPDGGADPVAGSWAPHLLSEVTASPAPTWDAPPDNASGCGEQINYGRVEKVVIGSILTLITLLTIAGNCLVVISVCFVKKLRQPSNYLIVSLALADLSVAVAVMPFVSVTDLIGGKWIFGHFFCNVFIAMDVMCCTASIMTLCVISIDRYLGITRPLTYPVRQNGKCMAKMILSVWLLSASITLPPLFGWAQNVNDDKVCLISQDFGYTIYSTAVAFYIPMSVMLFMYYQIYKAARKSAAKHKFPGFPRVEPDSVIALNGIVKLQKEVEECANLSRLLKHERKNISIFKREQKAATTLGIIVGAFTVCWLPFFLLSTARPFICGTSCSCIPLWVERTFLWLGYANSLINPFIYAFFNRDLRTTYRSLLQCQYRNINRKLSAAGMHEALKLAERPERPEFVLRACTRRVLLRPEKRPPVSVWVLQSPDHHNWLADKMLTTVEKKVMIHD | |
| 61202332333024302320134334313423443333323232333123333333233342323313332342423200000000000200220131020000000112300000000000000000000000001000000021101102100000000000000000100000000000000200303331122000000000111010001000000223273542020112210000000100310000002001100100132133222222232232233323333433343342232323222233222323443311310000000000001100000001000231323200100000001201200110000000104100300130010222434342434333432433344442333243444433234564343433434345333322431121134303138 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 440 460 | | | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCSSCCCCCSSSSCC MMDVNSSGRPDLYGHLRSFLLPEVGRGLPDLSPDGGADPVAGSWAPHLLSEVTASPAPTWDAPPDNASGCGEQINYGRVEKVVIGSILTLITLLTIAGNCLVVISVCFVKKLRQPSNYLIVSLALADLSVAVAVMPFVSVTDLIGGKWIFGHFFCNVFIAMDVMCCTASIMTLCVISIDRYLGITRPLTYPVRQNGKCMAKMILSVWLLSASITLPPLFGWAQNVNDDKVCLISQDFGYTIYSTAVAFYIPMSVMLFMYYQIYKAARKSAAKHKFPGFPRVEPDSVIALNGIVKLQKEVEECANLSRLLKHERKNISIFKREQKAATTLGIIVGAFTVCWLPFFLLSTARPFICGTSCSCIPLWVERTFLWLGYANSLINPFIYAFFNRDLRTTYRSLLQCQYRNINRKLSAAGMHEALKLAERPERPEFVLRACTRRVLLRPEKRPPVSVWVLQSPDHHNWLADKMLTTVEKKVMIHD | |||||||||||||||||||||||||
| 1 | 3sn6R | 0.36 | 0.22 | 0.58 | 2.06 | Download | -----------------------------------------------------------------------------EVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILT-KTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRYAEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQKIDKSEGR---------------------------------------CLKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQD----NLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLC------------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 4ib4A | 0.26 | 0.23 | 0.67 | 3.79 | Download | --------------------------------------------------------------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQEAQAAAEQLKTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQT-TLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------------------------- | |||||||||||||||||||
| 3 | 5uenA | 0.24 | 0.20 | 0.69 | 3.08 | Download | ---------------------------------------------------------------------------SISAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIG-PQTYF--HTCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLAGSMGEPVIISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLNVKDAAPEMKDFRHGFDILVGQIDDALKLANEAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPSCH---KPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPLEVLF------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.28 | 0.23 | 0.68 | 1.56 | Download | ----------------------------------------------------------------------EEQG----NKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVIKGIET-NPNNITCVLTKERDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNKADAALDAQKKRGGKKEAQEQLKTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSC-NQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------------------------- | |||||||||||||||||||
| 5 | 3uon | 0.30 | 0.23 | 0.66 | 1.22 | Download | -----------------------------------------------------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYT-VIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFITVEDGECYIQFNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKGAAAIGRNTNGVITKKAGAKTTFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAP----CIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 5uenA | 0.23 | 0.20 | 0.69 | 2.64 | Download | -----------------------------------------------------------------------------SAFQAAYIGIEVLIALVSVPGNVLVIWAVKVNQALRDATFCFIVSLAVADVAVGALVIPLAILINIG-PQTYFH--TCLMVACPVLILTQSSILALLAIAVDRYLRVKIPLRYKMVVTPRRAAVAIAGCWILSFVVGLTPMFGWNNLSAVEEFEKVISMEYMVYFNFFVWVLPPLLLMVLIYLEVFYLIRKQLADLEDNWETLNDNVKDALTKMEAQAAAEQLKTTRNAYIQKYLERARSTLQKELKIAKSLALILFLFALSWLPLHILNCITLFCPS---CHKPSILTYIAIFLTHGNSAMNPIVYAFRIQKFRVTFLKIWNDHFRCQPLEVLF------------------------------------------------------------------- | |||||||||||||||||||
| 7 | 4ib4 | 0.28 | 0.23 | 0.67 | 1.75 | Download | --------------------------------------------------------------------------------LHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPPIKGIETN-PNNITCVLTKEGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNKADNAARAALDAQKGVKEAQAQTTRNAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.31 | 0.25 | 0.67 | 4.28 | Download | ----------------------------------------------------------------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKM-WTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRATEETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLQMGETGVAGFTNSLRMLQQKRWDEAAVNLAKSRWYNQTPNRAKRVITTFRTGHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQD----NLIRKEVYILLNWIGYVNSGFNPLIYC-RSPDFRIAFQELLCL------------------------------------------------------------------------------ | |||||||||||||||||||
| 9 | 4ib4A | 0.26 | 0.23 | 0.69 | 2.56 | Download | --------------------------------------------------------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIETNPNNITLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCN-QTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 3zpqA | 0.37 | 0.24 | 0.58 | 3.10 | Download | -----------------------------------------------------------------------GAELLSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVR-GTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQS---------------------------------------------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRD----LVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRLLA------------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
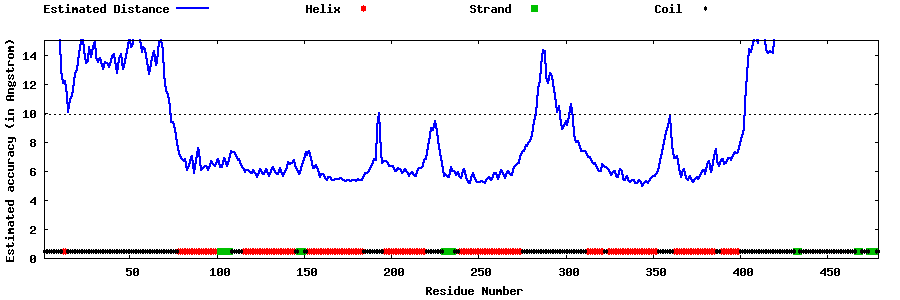
| Generated 3D models | Estimated local accuracy of models | ||
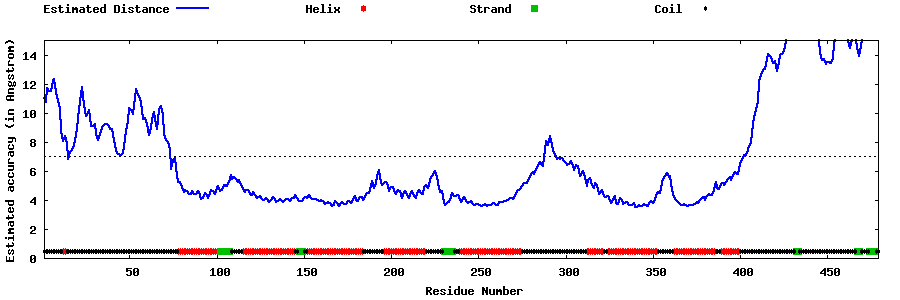 | |||
|
|
|||
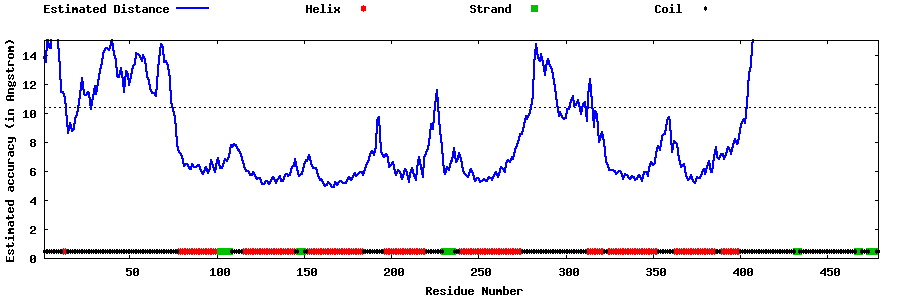 | |||
|
| |||
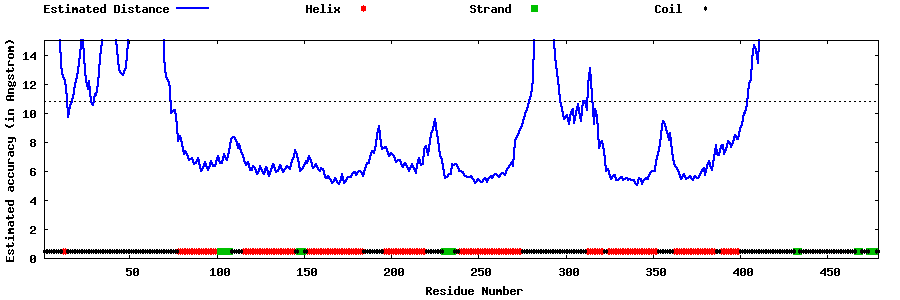 | |||
|
| |||
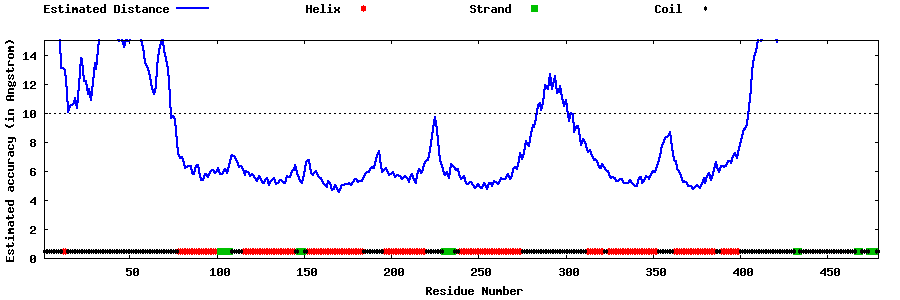 | |||
|
| |||
 | |||
|
| |||