

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MVWGKICWFSQRAGWTVFAESQISLSCSLCLHSGDQEAQNPNLVSQLCGVFLQNETNETIHMQMSMAVGQQALPLNIIAPKAVLVSLCGVLLNGTVFWLLCCGATNPYMVYILHLVAADVIYLCCSAVGFLQVTLLTYHGVVFFIPDFLAILSPFSFEVCLCLLVAISTERCVCVLFPIWYRCHRPKYTSNVVCTLIWGLPFCINIVKSLFLTYWKHVKACVIFLKLSGLFHAILSLVMCVSSLTLLIRFLCCSQQQKATRVYAVVQISAPMFLLWALPLSVAPLITDFKMFVTTSYLISLFLIINSSANPIIYFFVGSLRKKRLKESLRVILQRALADKPEVGRNKKAAGIDPMEQPHSTQHVENLLPREHRVDVET | |
| CCCCCSSSSCCCCCCSSCCCCCCCCCCSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 965220210268997041455234631123016887889976555555663247888888988765553024689999999999999999899999999985556466999999999999999987999999993499986317999999999999999999999999998898873776156762488999999999999999978999723445875137750788899999999999999999999998367888864788877887788999689999999999999999999999999999999999999369978999999999999986637666678998998994777787433566799853113579 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MVWGKICWFSQRAGWTVFAESQISLSCSLCLHSGDQEAQNPNLVSQLCGVFLQNETNETIHMQMSMAVGQQALPLNIIAPKAVLVSLCGVLLNGTVFWLLCCGATNPYMVYILHLVAADVIYLCCSAVGFLQVTLLTYHGVVFFIPDFLAILSPFSFEVCLCLLVAISTERCVCVLFPIWYRCHRPKYTSNVVCTLIWGLPFCINIVKSLFLTYWKHVKACVIFLKLSGLFHAILSLVMCVSSLTLLIRFLCCSQQQKATRVYAVVQISAPMFLLWALPLSVAPLITDFKMFVTTSYLISLFLIINSSANPIIYFFVGSLRKKRLKESLRVILQRALADKPEVGRNKKAAGIDPMEQPHSTQHVENLLPREHRVDVET | |
| 730030000143242312242313232332233442333334324333333331312313334334323332021111222022002312321020000000233200000000000000000000001000001332111010001000001330121001000000000000000000012212310000000000220010010000000023332010001002333332223032201000000001224434431000000000100000122110000021022032011001000002002200000000340152014002200320042345446665444444454544456355443464465468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCSSSSCCCCCCSSCCCCCCCCCCSSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MVWGKICWFSQRAGWTVFAESQISLSCSLCLHSGDQEAQNPNLVSQLCGVFLQNETNETIHMQMSMAVGQQALPLNIIAPKAVLVSLCGVLLNGTVFWLLCCGATNPYMVYILHLVAADVIYLCCSAVGFLQVTLLTYHGVVFFIPDFLAILSPFSFEVCLCLLVAISTERCVCVLFPIWYRCHRPKYTSNVVCTLIWGLPFCINIVKSLFLTYWKHVKACVIFLKLSGLFHAILSLVMCVSSLTLLIRFLCCSQQQKATRVYAVVQISAPMFLLWALPLSVAPLITDFKMFVTTSYLISLFLIINSSANPIIYFFVGSLRKKRLKESLRVILQRALADKPEVGRNKKAAGIDPMEQPHSTQHVENLLPREHRVDVET | |||||||||||||||||||||||||
| 1 | 4n6hA | 0.14 | 0.21 | 0.87 | 2.16 | Download | TKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGNEGKVKEAQAAAEQLKTTRNAYIQKYLGSPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYMKTATNIYIFNLALADALATSTLPFQSAKYLM-ETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK----------------------------------------------- | |||||||||||||||||||
| 2 | 4bwbA | 0.13 | 0.16 | 0.71 | 4.03 | Download | --------------------------------------------------------------NSDLDVNTDIYSKVLVTAIYLALFVVGTVGNGVTLFTLAQSLQSRVDYYLGSLALSDLLILLALPVDLYNFIVHHPWAFGDAGCKGYYFLREACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHVIQLNTFMSFLFPMLVASILNTVAARRLTVMVEPGRRRGVLVLRAVVIAFVVCWLPYHVRRLMFVYDFYHYFYMLSNALVYVSAAINPILYNLVSANFRQVFLSTLAC----------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.15 | 0.18 | 0.72 | 3.58 | Download | --------------------------------------------------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRKMKTATNIYIFNLALADALATSTLPFQSAKYLMET-WPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLLSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG-------------------------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.17 | 0.16 | 0.69 | 1.53 | Download | ----------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIITKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPF-GDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVDVIECSLMICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSYYFCIALGYTNSSLNPILYAFLD----ENFKRCFRDFCFP------------------------------------------- | |||||||||||||||||||
| 5 | 4djh | 0.16 | 0.16 | 0.69 | 1.16 | Download | ----------------------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVDVIECSKICVFIFAFVIPVLIIIVCYTLMILRLKSVRDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------------------------------- | |||||||||||||||||||
| 6 | 5o9hA | 0.21 | 0.20 | 0.72 | 2.82 | Download | ---------------------------------------------------------------------TLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEARTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYRRERAVAIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMSFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------------------------------------- | |||||||||||||||||||
| 7 | 4djh | 0.17 | 0.16 | 0.67 | 1.70 | Download | ---------------------------------------------------------------------------VIITAVYSVVFVVGLVGNSLVMFVIIRKMKTATNIYIFNLALADALVTTTMPFQSTV-YLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRVIECSLQVFIFAFVIPVLIIIVCYTLMILRLKSVRLLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFC------------------------------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.16 | 0.16 | 0.70 | 3.29 | Download | ---------------------------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHKIVILGLVLPLLVMVICYSGILKTLLRE--KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ------------------------------------------------ | |||||||||||||||||||
| 9 | 5o9hA | 0.21 | 0.20 | 0.72 | 2.61 | Download | --------------------------------------------------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVL-IVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKVVVAVVASFFIFWLPYQVTGIMMSFPTFLLLKKLDSLCVSFAYINCCINPIIYVVAGQGFQKSLPELLREVLTEESVVR-------------------------------------- | |||||||||||||||||||
| 10 | 5o9hA | 0.21 | 0.20 | 0.72 | 2.88 | Download | --------------------------------------------------------------------NTLRVPDILALVIFAVVFLVGVLGNALVVWVTAFEKRTINAIWFLNLAVADFLACLALPALFTSIVQHHHWPFGGAACSILPSLILLNMYASILLLATISADRFLLVFKPAWCQRFRGAGLAWILCAVAWGLALLLTIPSALYRVVREEYFPPKVLCIVRLVLGFLWPLLTLTICYTFILLRTWSARETRSTKTLKVVVAVVASFFIFWLPYQVTGIMMPTFLLLKLDSLCVSFAYINCCINPIIYVVAGQGFQK----SLPELLREVLTEESVVR---------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
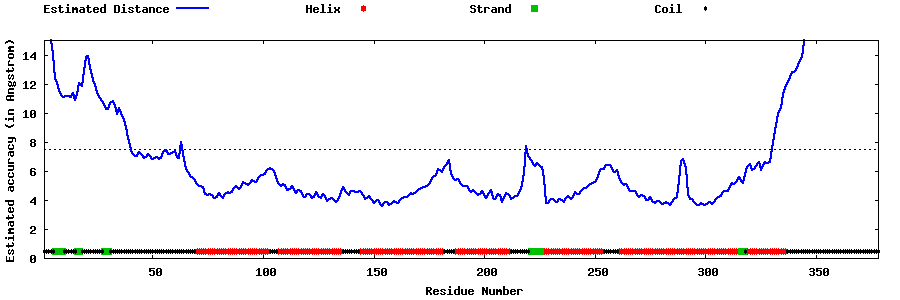 | |||
|
|
|||
 | |||
|
| |||
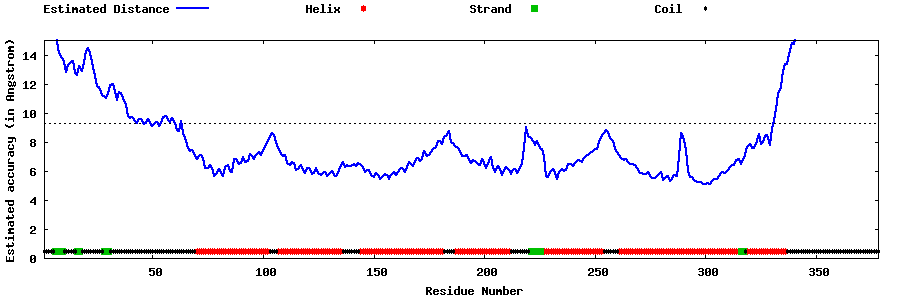 | |||
|
| |||
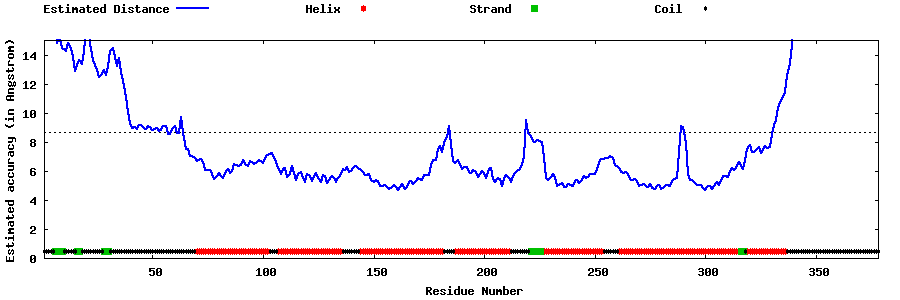 | |||
|
| |||
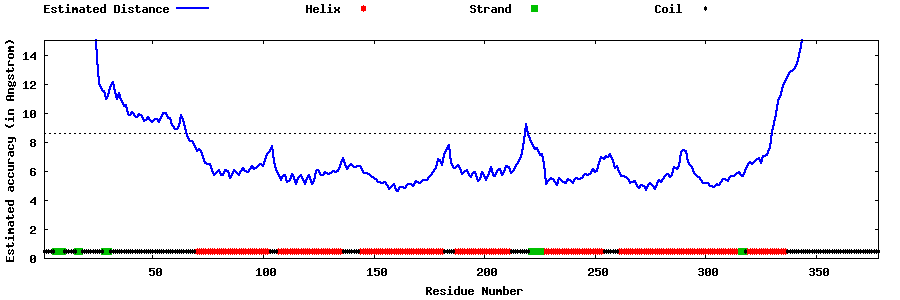 | |||
|
| |||