

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MDSGPLWDANPTPRGTLSAPNATTPWLGRDEELAKVEIGVLATVLVLATGGNLAVLLTLGQLGRKRSRMHLFVLHLALTDLAVALFQVLPQLLWDITYRFQGPDLLCRAVKYLQVLSMFASTYMLLAMTLDRYLAVCHPLRSLQQPGQSTYLLIAAPWLLAAIFSLPQVFIFSLREVIQGSGVLDCWADFGFPWGPRAYLTWTTLAIFVLPVTMLTACYSLICHEICKNLKVKTQAWRVGGGGWRTWDRPSPSTLAATTRGLPSRVSSINTISRAKIRTVKMTFVIVLAYIACWAPFFSVQMWSVWDKNAPDEDSTNVAFTISMLLGNLNSCCNPWIYMGFNSHLLPRPLRHLACCGGPQPRMRRRLSDGSLSSRHTTLLTRSSCPATLSLSLSLTLSGRPRPEESPRDLELADGEGTAETIIF | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9999988899998878788888887678771399999999999999999999998898742899884648999999999999999988999999983847171588879999999999999999999999858984453876574088899999999999999999999986899835998369982788870044589898889999999999999999999999885110000110133332224577765322124565444320347888777765546589999999817999999999973456653428999999999999999999999993499999999997456688987656677888777677876777899998877767777999899899876778887766543469 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | |
| | | | | | | | | | | | | | | | | | | | | | | |
| MDSGPLWDANPTPRGTLSAPNATTPWLGRDEELAKVEIGVLATVLVLATGGNLAVLLTLGQLGRKRSRMHLFVLHLALTDLAVALFQVLPQLLWDITYRFQGPDLLCRAVKYLQVLSMFASTYMLLAMTLDRYLAVCHPLRSLQQPGQSTYLLIAAPWLLAAIFSLPQVFIFSLREVIQGSGVLDCWADFGFPWGPRAYLTWTTLAIFVLPVTMLTACYSLICHEICKNLKVKTQAWRVGGGGWRTWDRPSPSTLAATTRGLPSRVSSINTISRAKIRTVKMTFVIVLAYIACWAPFFSVQMWSVWDKNAPDEDSTNVAFTISMLLGNLNSCCNPWIYMGFNSHLLPRPLRHLACCGGPQPRMRRRLSDGSLSSRHTTLLTRSSCPATLSLSLSLTLSGRPRPEESPRDLELADGEGTAETIIF | |
| 6463442221313432333333333323232200100021102002101200000000001233120000000100020000000000101000001332100100010030010000000000000002330100000020232133000000000010000000100000002323644200000010234202000000002220330010002000000010133132444434344443433434444444444444443344443234122100000000000000002010000001001330243301100000001000200000000000004400420141020023344345444443333333233334344444444434333454444663454342363643333144 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 420 | | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSSSCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MDSGPLWDANPTPRGTLSAPNATTPWLGRDEELAKVEIGVLATVLVLATGGNLAVLLTLGQLGRKRSRMHLFVLHLALTDLAVALFQVLPQLLWDITYRFQGPDLLCRAVKYLQVLSMFASTYMLLAMTLDRYLAVCHPLRSLQQPGQSTYLLIAAPWLLAAIFSLPQVFIFSLREVIQGSGVLDCWADFGFPWGPRAYLTWTTLAIFVLPVTMLTACYSLICHEICKNLKVKTQAWRVGGGGWRTWDRPSPSTLAATTRGLPSRVSSINTISRAKIRTVKMTFVIVLAYIACWAPFFSVQMWSVWDKNAPDEDSTNVAFTISMLLGNLNSCCNPWIYMGFNSHLLPRPLRHLACCGGPQPRMRRRLSDGSLSSRHTTLLTRSSCPATLSLSLSLTLSGRPRPEESPRDLELADGEGTAETIIF | |||||||||||||||||||||||||
| 1 | 5vblB | 0.19 | 0.21 | 0.79 | 2.48 | Download | ------------------------CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTN-KVQCYMDYSMVAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGNPGTDFKDIPDDWVCPLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR----------------------------------------------------------------- | |||||||||||||||||||
| 2 | 5gliA | 0.20 | 0.21 | 0.79 | 3.85 | Download | ------------------SPPPCQGPIEIKETFKYINTVVSCLVFVLGIIGNSTLLYIIYKNKCMRNGPNILIASLALGDLLHIVIAIPINVYKLLAEDWPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVASWSRIKGGVPKWTAVEIVLIWVVSVVLAVPEAIGFDIITMYKGSYLRICLLHPVQKFYATAKDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNIFEMLRIDEGGGSGGDEAEKLFNQDVDAAVRTGTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKLTLYNQNDPNRLLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSALCC--------------------------------------------------------------------- | |||||||||||||||||||
| 3 | 4n6hA | 0.22 | 0.23 | 0.71 | 3.47 | Download | ---------------------SPGARSASSLALAIAITALYSAVCAVGLLGNVLVMFGIVRYTKMKTATNIYIFNLALADALATST-LPFQSAKYLMETWPFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDG---AVVCMLQFPSPYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSVRLL----------------------------------SGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIDRRDPLVVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRKPCG------------------------------------------------------------------ | |||||||||||||||||||
| 4 | 4djh | 0.19 | 0.19 | 0.75 | 1.55 | Download | -----------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTT-MPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRPLKAKIINICIWLLSSSVGISAIVLGGTKVR-EDVDVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFEIDEGLRTGTGRNTTKKLNSLQQKTPGTWDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAA------LSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--------------------------------------------------------------------- | |||||||||||||||||||
| 5 | 4rnb | 0.23 | 0.20 | 0.76 | 1.17 | Download | -----------------------------PKEYEWVLIAGYIIVFVVALIGNVLVCVAVWKNHHMRTVTNYFIVNLSLADVLVTITCLPATLVVDITETWFFGQSLCKVIPYLQTVSVSVSVLTLSCIALDRWYAICHPST-----AKRARNSIVIIWIVSCIIMIPQAIVMECSTVFKTTLFTVCDERWGGEIYPKMYHICFFLVTYMAPLCLMVLAYLQIFRKLWCRQGIDCSFWNESYLTDRFRGQKGKGDTNETGRSLKKRAMSFSKQIRARRKTARMLMVVLLVFAICYLPISILNVLKRVFGMFADRETVYAWFTFSHWLVYANSAANPIIYNFLSGKFREEFKAAFSC--------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 5vblB | 0.20 | 0.21 | 0.78 | 2.84 | Download | --------------------------YTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSSRKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARRLRVSGAVATAVLWVLAALLAMPVMVLRTTGDLENTN-KVQCYMDYSEWAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGDPDNGVNPGTDFKDIPLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWDLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR----------------------------------------------------------------- | |||||||||||||||||||
| 7 | 3uon | 0.20 | 0.22 | 0.74 | 1.68 | Download | ----------------------------------VFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFF---SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFEMLRIDEGLRLKAAGRNTVITKGNAKAGFRTGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCI----PNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4bvnA | 0.22 | 0.19 | 0.68 | 4.39 | Download | ----------------------------LSQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQIRKRKTS---------------------------------RVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP----KWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA---------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 5vblB | 0.19 | 0.21 | 0.79 | 2.78 | Download | ------------------------CEYTDWKSSGALIPAIYMLVFLLGTTGNGLVLWTVFRSREKRRSADIFIASLAVADLTFVVTLPLWATYTYRDYDWPFGTFFCKLSSYLIFVNMYASAFCLTGLSFDRYLAIVRPVANARLLRVSGAVATAVLWVLAALLAMPVMVLRTTGD-LENTNKVQCYMDYSMVAWEVGLGVSSTTVGFVVPFTIMLTCYFFIAQTIAMKKYTCTVCGYIYNPEDGNPGTDFKDIPDDWVCPLCGVGKDQFEEVEERRRLLSIIVVLVVTFALCKMPYHLVKTLYMLGSLLHWPLFLMNIFPYCTCISYVNSCLNPFLYAFFDPRFRQACTSMLLMGQSR----------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.22 | 0.20 | 0.69 | 2.54 | Download | ------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALAT-STLPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQG---SIDCTLTFSHWYWENLLKICVFIFAFIMPVLIITVCYGLMILRLK----------------------------------SVRMLSGSKEKDRNLRRITRMVLVVVAVFIVCWTPIHIYVIIKALI-TIPETTFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF--------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
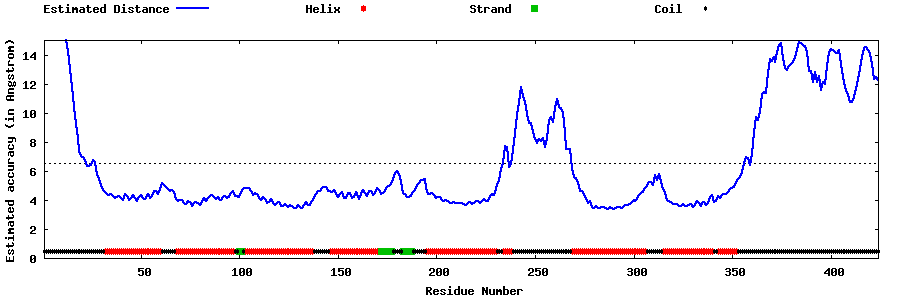 | |||
|
|
|||
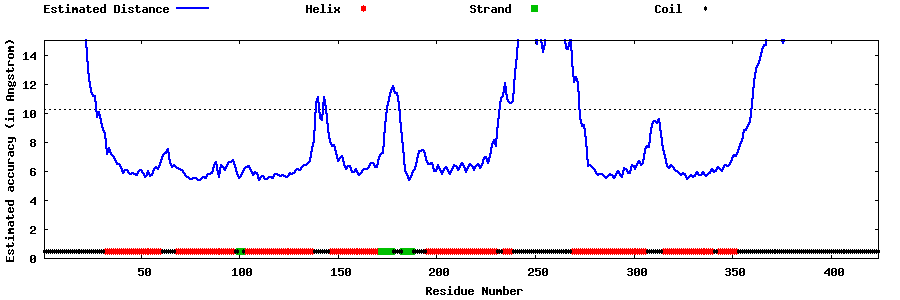 | |||
|
| |||
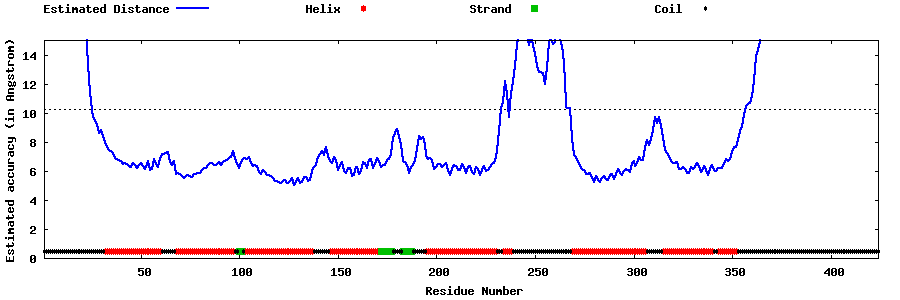 | |||
|
| |||
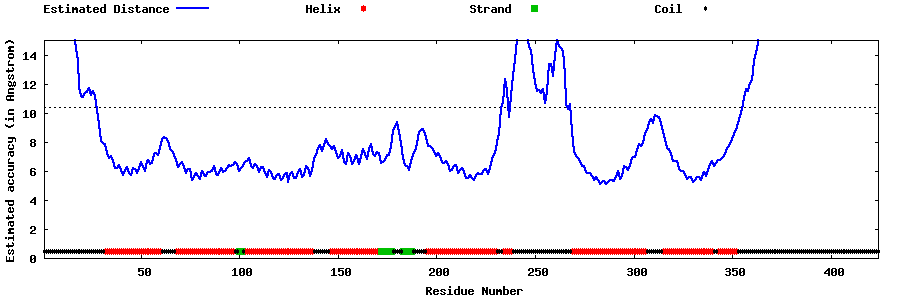 | |||
|
| |||
 | |||
|
| |||