

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MASPRLGTFCCPTRDAATQLVLSFQPRAFHALCLGSGGLRLALGLLQLLPGRRPAGPGSPATSPPASVRILRAAAACDLLGCLGMVIRSTVWLGFPNFVDSVSDMNHTEIWPAAFCVGSAMWIQLLYSACFWWLFCYAVDAYLVIRRSAGLSTILLYHIMAWGLATLLCVEGAAMLYYPSVSRCERGLDHAIPHYVTMYLPLLLVLVANPILFQKTVTAVASLLKGRQGIYTENERRMGAVIKIRFFKIMLVLIICWLSNIINESLLFYLEMQTDINGGSLKPVRTAAKTTWFIMGILNPAQGFLLSLAFYGWTGCSLGFQSPRKEIQWESLTTSAAEGAHPSPLMPHENPASGKVSQVGGQTSDEALSMLSEGSDASTIEIHTASESCNKNEGDPALPTHGDL | |
| CCCCCCCCSSCCCCCHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCSSHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 99977675436877734566553040477743499999999999998877765056654200344234899999999999999999999999988888753553057767787763899999999999999999999999986146656540311103456388899999988740367765210146422899999999999999999999999999999999875043257889999999999999999999999999999999876401211012200000027999999999999999999999999876344763203212214576777778857899878889999876788888894443046999887755045664445756689898254779 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | |
| | | | | | | | | | | | | | | | | | | | | | |
| MASPRLGTFCCPTRDAATQLVLSFQPRAFHALCLGSGGLRLALGLLQLLPGRRPAGPGSPATSPPASVRILRAAAACDLLGCLGMVIRSTVWLGFPNFVDSVSDMNHTEIWPAAFCVGSAMWIQLLYSACFWWLFCYAVDAYLVIRRSAGLSTILLYHIMAWGLATLLCVEGAAMLYYPSVSRCERGLDHAIPHYVTMYLPLLLVLVANPILFQKTVTAVASLLKGRQGIYTENERRMGAVIKIRFFKIMLVLIICWLSNIINESLLFYLEMQTDINGGSLKPVRTAAKTTWFIMGILNPAQGFLLSLAFYGWTGCSLGFQSPRKEIQWESLTTSAAEGAHPSPLMPHENPASGKVSQVGGQTSDEALSMLSEGSDASTIEIHTASESCNKNEGDPALPTHGDL | |
| 72424242000223342132234022300000000000000000000000222344333333133010000000000110001011020001000120031023133232221000100000000001000000000031000000223433101000000012000000200001103333102001210001111132122200210110000001101231343344345532430230010001003003300200010101212232333133332332320100000000100031000000001002422341424434241543433444443434333343443443434444434431433446364334413443443447434424554355 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 400 | | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCSSCCCCCHHHHHHHHCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCSSHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MASPRLGTFCCPTRDAATQLVLSFQPRAFHALCLGSGGLRLALGLLQLLPGRRPAGPGSPATSPPASVRILRAAAACDLLGCLGMVIRSTVWLGFPNFVDSVSDMNHTEIWPAAFCVGSAMWIQLLYSACFWWLFCYAVDAYLVIRRSAGLSTILLYHIMAWGLATLLCVEGAAMLYYPSVSRCERGLDHAIPHYVTMYLPLLLVLVANPILFQKTVTAVASLLKGRQGIYTENERRMGAVIKIRFFKIMLVLIICWLSNIINESLLFYLEMQTDINGGSLKPVRTAAKTTWFIMGILNPAQGFLLSLAFYGWTGCSLGFQSPRKEIQWESLTTSAAEGAHPSPLMPHENPASGKVSQVGGQTSDEALSMLSEGSDASTIEIHTASESCNKNEGDPALPTHGDL | |||||||||||||||||||||||||
| 1 | 4l6rA | 0.14 | 0.18 | 0.70 | 1.45 | Download | -------------EVQKEVAKMYSSFQVMYTVGYSLSLGALLLALAILGGLSKLHC---------TRNAIHANLFASFVLKASSVLVIDGLLR-------------TLSDGAVAGCRVAAVFMQYGIVANYCWLLVEGLYLHNLLGLLPERSFFSLYLGIGWGAPMLFVVPWAVVKCLFENVQCWTSNDN-MGFWWILRFPVFLAILINFFIFVRIVQLLVAKLRARQMHHT----DYKFRLAKSTLTLIPLLGVHEVVFAF---------------VTDEHAQGTLRSAKLFFDLFLSSFQGLLVAVLYCFLNKEVQSELRRRWHRWRLGKVLWEERN----------------------------------------------------------------- | |||||||||||||||||||
| 2 | 5l7dA | 0.09 | 0.21 | 0.90 | 2.16 | Download | PPPLSHCGIQCQNPLFTEAEHQDMHSYIAAFGAVTGLCTLFTLATFVADWRNSNRYP--------AVILFYVNACFFVGSIGWLAQFMDGARREIVCRADGTMRLGEPTSNETLSCVIIFVIVYYALMAGFVWFVVLTYAWHTSFKALQPSGKTSYFHLLTWSLPFVLTVAILAVAQDSVSGICFVGYKNYRYRAGFVLAPIGLVLIVGGYFLIRGVMTLFSARRQLAD--LEDNWETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYIQKYLERARSTLSKINVLCQANVTIGLPTKQPIPDCEIKNRPSLLVEKIT-------------------------------- | |||||||||||||||||||
| 3 | 4k5yA | 0.14 | 0.17 | 0.61 | 2.78 | Download | --------------------------HVAAIINYLGHCISLVALLVAFVLFLR------ARSIRCLRNIIHANLIAAFILRNATWFVVQLTMSPEVH------------QSNVGWCRLVTAAYNYFHVTNFFWMFGEGCYLHTAIV-LTDRLRAWMFICIGWGVPFPIIVAWAIGKLYYDNEKCWAGKRPGVYTDYIYQGPMALVLLINFIFLFNIVRILMTKL----RASTTSETIQARKAVKATLVLLPLLGITYMLAFV-----------------NPGEDEVSRVVFIYFNAFLESFQGFFVSVFACFLN------------------------------------------------------------------------------------------ | |||||||||||||||||||
| 4 | 5ee7A | 0.15 | 0.13 | 0.64 | 1.80 | Download | ----------------------YSSFQVMYTVGYSLSLAALLLALAILGGLSKLHCT---------ANAIHANLFLSFVLKASAVLFIDGLLR---------TVSTWLSDGAVAACRVAAVFMQYGIVANYCWLLVEGLYLHNLLG---LRSFFSLYLGIGWGAPALFVVPWAVVKCLFENVQCW----TNMGFWWILRFPVFLAILINFFIFVRIVQLLVAKLRARQMHHTD----YAFRLAKSTLTLIPLLGVHFVVFAFVTD------------EHR--------SAKLFFDLALSSFQGLLVAVLYCFLNKEVQSELRRRWHRA---------------------------------------------------------------------------- | |||||||||||||||||||
| 5 | 4ib4 | 0.13 | 0.16 | 0.70 | 1.51 | Download | ---------------EE----QGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQ------YATNYFLMSLAVADLLVGLFVMPIALLTIMFEAMWP----------LPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQAATAFIKITVVWLISIGIAIPVPIKGIETNNITCVLTKERGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDKYLQTISNEQRASK-VLGIVFFLFLLMWCPFFITNITLVLCDSC--------N--QTTLQMLLEIFVWIGYVSSGVNPLV-YTLFNKTFRDAFGRYITCNYR------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4k5y | 0.15 | 0.17 | 0.61 | 2.64 | Download | ------------------------HYHVAAIINYLGHCISLVALLVAFVLFLRARSI------RCLRNIIHANLIAAFILRNATWFVVQL------------TMSPEVHQSNVGWCRLVTAAYNYFHVTNFFWMFGEGCYLHTAIVLTYDRLRAWMFICIGWGVPFPIIVAWAIGKLYYDNEKCWAGKRPGVYTDYIYQGPMALVLLINFIFLFNIVRILMTKLRAS----TTSETIQARKAVKATLVLLPLLGITYMLAFVN---------------------EVSRVVFIYFNAFLESFQGFFVSVFACFLNS----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 7 | 4l6rA | 0.14 | 0.18 | 0.70 | 1.98 | Download | ---------------QKEVAKMYSSFQVMYTVGYSLSLGALLLALAILGGLSKLHC---------TRNAIHANLFASFVLKASSVLVIDGLLR-------------TLSDGAVAGCRVAAVFMQYGIVANYCWLLVEGLYLHNLLGLLPERSFFSLYLGIGWGAPMLFVVPWAVVKCLFENVQCW-TSNDNMGFWWILRFPVFLAILINFFIFVRIVQLLVAKLRARQM----HHTDYKFRLAKSTLTLIPLLGVHEVVFAF---------------VTDEHAQGTLRSAKLFFDLFLSSFQGLLVAVLYCFLNKEVQSELRRRWHRWRLGKVLWEERN----------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4ib4 | 0.13 | 0.16 | 0.65 | 1.67 | Download | ------------------------------LMVIIPTIGGNTLVILAVSLE---K------KLQYATNYFLMSLAVADLLVGLFVMPIALLTIM----------FEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANTAFIKITVVWLISIGIAIPVPIKGIETNPNNITCVLTKERFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCD---SCNQTTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGR---------------------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 4k5yA | 0.15 | 0.15 | 0.59 | 2.02 | Download | -----------------------------HYLGHCISLVALLVAFVLFLRARSIRC---------LRNIIHANLIAAFILRNATWFVVQLTMS------------PEVHQSNVGWCRLVTAAYNYFHVTNFFWMFGEGCYLHTAIVLT-DRLRAWMFICIGWGVPFPIIVAWAIGKLYYDNEKCWAGKRPGVYTDYIYQGPMALVLLINFIFLFNIVRILMTK----LRASTTSETIQARKAVKATLVLLPLLGITYMLAFVN---------------------EVSRVVFIYFNAFLESFQGFFVSVFACFLNS----------------------------------------------------------------------------------------- | |||||||||||||||||||
| 10 | 5vaiR | 0.13 | 0.15 | 0.70 | 1.74 | Download | -------------SSPEERLLSL---YIIYTVGYALSFSALVIASAILLGFRHLHCT---------RNYIHLNLFASFILRALSVFIKDAALKWMYSTAAQHQWDGLLSYQDSLGCRLVFLLMQYCVAANYYWLLVEGAYLYTLLAFASEQRIFKLYLSIGWGVPLLFVIPWGIVKYLYEDEGCWTRNSNM-NYWLIIRLPILFAIGVNFLIFIRVICIVVSKLKANL----MCKTDIKCRLAKSTLTLIPLLGTHEVIFAFV---------------MDEHARGTLRFVKLFTELSFTSFQGLMVAILYCFVNNEVQMEFRKSWERWR--------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
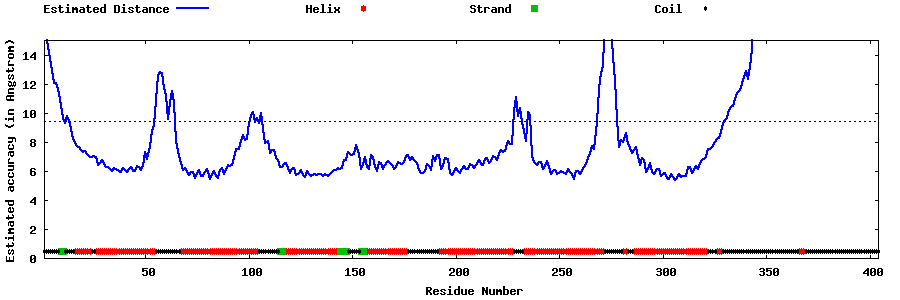
| Generated 3D models | Estimated local accuracy of models | ||
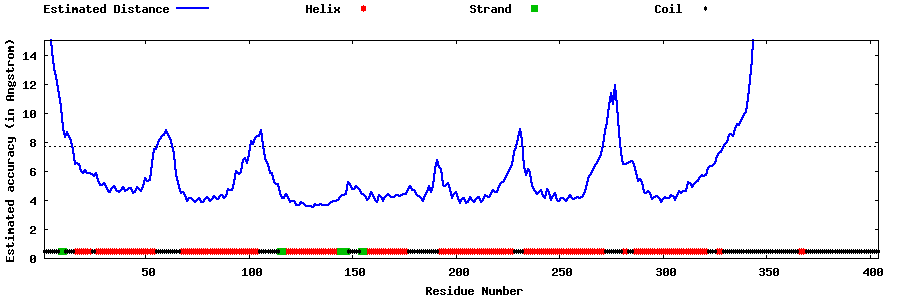 | |||
|
|
|||
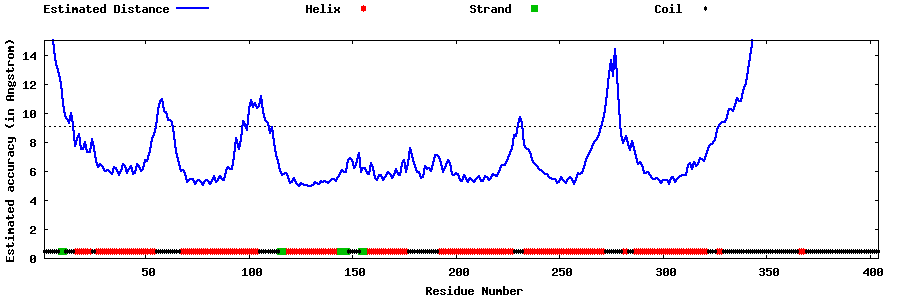 | |||
|
| |||
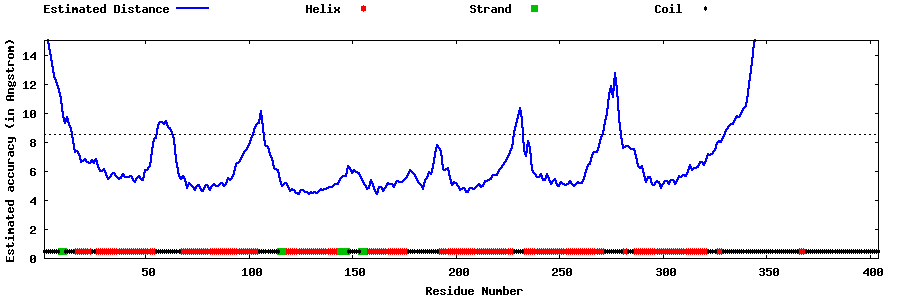 | |||
|
| |||
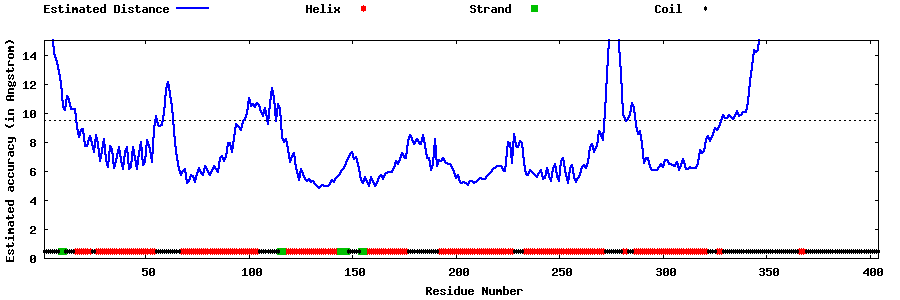 | |||
|
| |||
 | |||
|
| |||