

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MLNTTSVTEFLLLGVTDIQELQPFLFVVFLTIYFISVTGNGAVLMIVISDPRLHSLMYFFLGNLSYLDICYSTVTLPKMLQNFLSTHKAISFLGCISQLHFFHSLGSTESMLFAVMAFDLSVAICKPLRYTVIMNPQLCTQMAITIWVIGFFHALLHSVMTSRLNFCGSNRIHHFLCDIKPLLKLACGNTELNQWLLSTVTGTIAMGPFFLTLLSYFYIITYLFFKTRSCSMLCKALSTCASHFMVVILFYAPVLFTYIHPALESFMDQDRIVAIMYTVVTPVLNPLIYTLRNKEVKGALGRVIRRL | |
| CCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHCCHHHHHHHHHHHHCCSSSSSSCCCCCCCCCCCSSSSSSHHCCCCCCCCHHHCCCCHHHHHHHHHHHHHC | |
| 9988732533032699885079999999999999999989999999971788677499997889987441341588999898625996784999999999999999999999999987617761655447623588699999999999999999999999824179999946895668689988845464698999999999999998999999999999998020656366771003648579959997533036893789988788881899852050102245631256299999999998629 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MLNTTSVTEFLLLGVTDIQELQPFLFVVFLTIYFISVTGNGAVLMIVISDPRLHSLMYFFLGNLSYLDICYSTVTLPKMLQNFLSTHKAISFLGCISQLHFFHSLGSTESMLFAVMAFDLSVAICKPLRYTVIMNPQLCTQMAITIWVIGFFHALLHSVMTSRLNFCGSNRIHHFLCDIKPLLKLACGNTELNQWLLSTVTGTIAMGPFFLTLLSYFYIITYLFFKTRSCSMLCKALSTCASHFMVVILFYAPVLFTYIHPALESFMDQDRIVAIMYTVVTPVLNPLIYTLRNKEVKGALGRVIRRL | |
| 8724131110000000433611100023023313313332320020010003000000000000001000010031020010111543102020000011000100220010001001100000020020103013300000001012101300310010000020036140200001032003000110320021012212331331230033022201100201031053231000000001000001120010000102143334342000011023203310300002032014003300534 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHCCHHHHHHHHHHHHCCSSSSSSCCCCCCCCCCCSSSSSSHHCCCCCCCCHHHCCCCHHHHHHHHHHHHHC MLNTTSVTEFLLLGVTDIQELQPFLFVVFLTIYFISVTGNGAVLMIVISDPRLHSLMYFFLGNLSYLDICYSTVTLPKMLQNFLSTHKAISFLGCISQLHFFHSLGSTESMLFAVMAFDLSVAICKPLRYTVIMNPQLCTQMAITIWVIGFFHALLHSVMTSRLNFCGSNRIHHFLCDIKPLLKLACGNTELNQWLLSTVTGTIAMGPFFLTLLSYFYIITYLFFKTRSCSMLCKALSTCASHFMVVILFYAPVLFTYIHPALESFMDQDRIVAIMYTVVTPVLNPLIYTLRNKEVKGALGRVIRRL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.22 | 0.89 | 3.62 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAAR-RQLVHAAKSLAIIVGLFALCWLPLHIIN-CFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.22 | 0.89 | 2.21 | Download | -ENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSAPDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM---- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.18 | 0.85 | 2.21 | Download | ----------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH--------------------ILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH---LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK | |||||||||||||||||||
| 4 | 3uon | 0.15 | 0.21 | 0.90 | 1.58 | Download | ------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVG--VRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRAS-KSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.24 | 0.92 | 1.25 | Download | KTRNAYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKAL--K-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.22 | 0.89 | 3.86 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAA-RRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| 7 | 4iaq | 0.17 | 0.22 | 0.86 | 1.72 | Download | -------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF- | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.19 | 0.86 | 2.70 | Download | --NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGV------------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPRAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQ-------WTYVSAAINPILYNLVSANFRQVFLSTL--- | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.22 | 0.89 | 4.90 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAA-RRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.21 | 0.92 | 5.68 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARR-QLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| ||||||||||||||||||||||||||
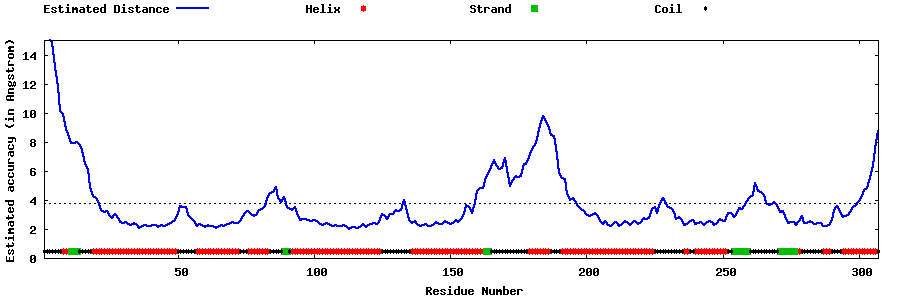
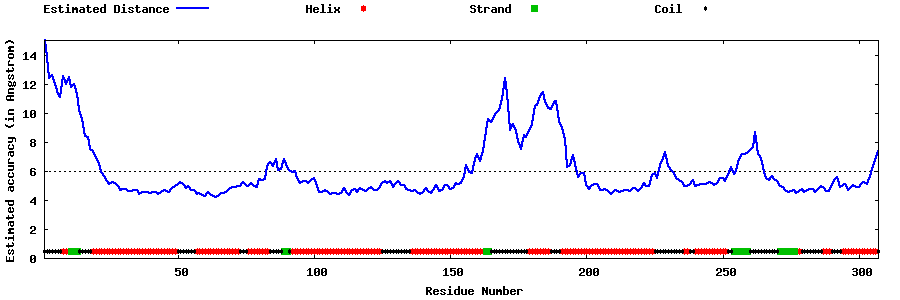
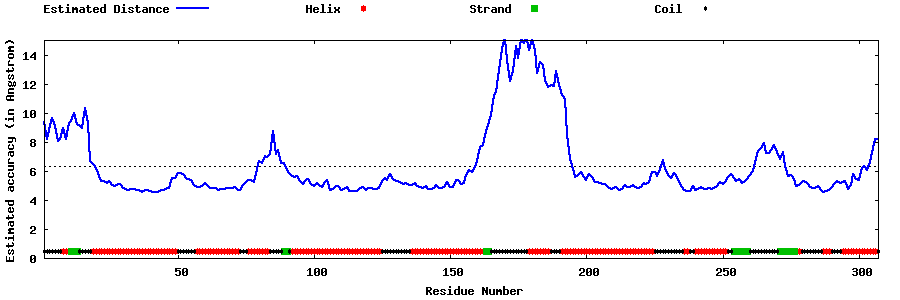
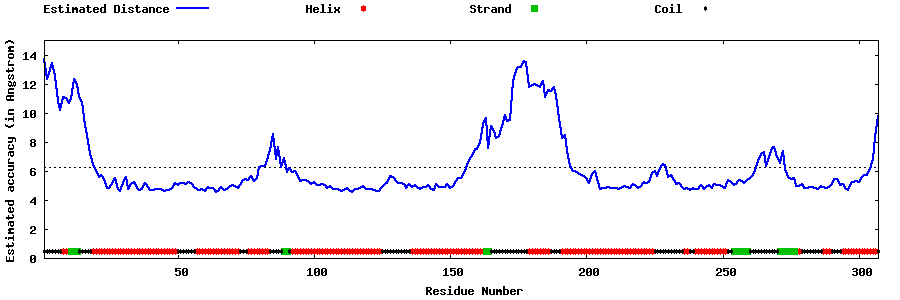
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||