

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MITFLPIIFSSLVVVTFVIGNFANGFIALVNSIEWFKRQKISFADQILTALAVSRVGLLWVLLLNWYSTVLNPAFNSVEVRTTAYNIWAVINHFSNWLATTLSIFYLLKIANFSNFIFLHLKRRVKSVILVMLLGPLLFLACHLFVINMNEIVRTKEFEGNMTWKIKLKSAMYFSNMTVTMVANLVPFTLTLLSFMLLICSLCKHLKKMQLHGKGSQDPSTKVHIKALQTVISFLLLCAIYFLSIMISVWSFGSLENKPVFMFCKAIRFSYPSIHPFILIWGNKKLKQTFLSVFWQMRYWVKGEKTSSP | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCSSCCCCCCCC | |
| 975899999999999999999999999999999998489888788999999999999999999727068757555662679889889999828999999999999706770899779999999656607899999999999999999613330047888833414561302688999999999999999999999999999999999863889999999819999999999999999999999999999998733762999999999999688988999419639999999999848123279556958 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MITFLPIIFSSLVVVTFVIGNFANGFIALVNSIEWFKRQKISFADQILTALAVSRVGLLWVLLLNWYSTVLNPAFNSVEVRTTAYNIWAVINHFSNWLATTLSIFYLLKIANFSNFIFLHLKRRVKSVILVMLLGPLLFLACHLFVINMNEIVRTKEFEGNMTWKIKLKSAMYFSNMTVTMVANLVPFTLTLLSFMLLICSLCKHLKKMQLHGKGSQDPSTKVHIKALQTVISFLLLCAIYFLSIMISVWSFGSLENKPVFMFCKAIRFSYPSIHPFILIWGNKKLKQTFLSVFWQMRYWVKGEKTSSP | |
| 733323321131133123313321220021001100443403201100000021100000113320000001120233300000101012212200100000010000000213110000012304210011022122323311131203221325434211123233332320111123333333233223323320210022024302433444411314002200100120221133113101311011233443202210200000112010001013143013001200210102036553668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCSSSCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCSSCCCCCCCC MITFLPIIFSSLVVVTFVIGNFANGFIALVNSIEWFKRQKISFADQILTALAVSRVGLLWVLLLNWYSTVLNPAFNSVEVRTTAYNIWAVINHFSNWLATTLSIFYLLKIANFSNFIFLHLKRRVKSVILVMLLGPLLFLACHLFVINMNEIVRTKEFEGNMTWKIKLKSAMYFSNMTVTMVANLVPFTLTLLSFMLLICSLCKHLKKMQLHGKGSQDPSTKVHIKALQTVISFLLLCAIYFLSIMISVWSFGSLENKPVFMFCKAIRFSYPSIHPFILIWGNKKLKQTFLSVFWQMRYWVKGEKTSSP | |||||||||||||||||||||||||
| 1 | 4djhA | 0.13 | 0.18 | 0.89 | 1.42 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSDR-----------NLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF-------------P | |||||||||||||||||||
| 2 | 5tjvA | 0.11 | 0.20 | 0.89 | 2.75 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPK----AVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIF-----PHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------- | |||||||||||||||||||
| 3 | 4djhA | 0.13 | 0.18 | 0.89 | 2.18 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMK-TATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKAL----DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL-------------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.12 | 0.22 | 0.92 | 1.56 | Download | EQGNKLHWAALLILMVIIPTIGGNTLVILAVSLE---KKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFEMWPLPLVLCAWLFLDVLFSTASIWHLCAISVDRYIAIK---KPIQAN-QYNSRATAFIKITVVWLIIGIAIPVPIGIETNPNNTCVLTKE-----RFGDFML--FGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLDNLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR---------- | |||||||||||||||||||
| 5 | 3odu | 0.15 | 0.14 | 0.96 | 1.27 | Download | NANFNKIFLPTIYSIIFLTGIVGNGLVILVMGY---QKKLRSMTDKYRLHLSVADLLFVIT-LPFWAVDAVANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYLAIVHATNSQRP----RKLLAEKVVYVGVWIPALLLTIPDFIFANVSEADDRYICDRFYPNDLWVVVFQFQHIMVGLILPGIVILSCYCIIISKLSHSGSNIFEMLRDAYGSKGHQKRKALKTTVILILAFFACWLPYYIGISIDFILLVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRPLEV---LFQ- | |||||||||||||||||||
| 6 | 4djhA | 0.12 | 0.18 | 0.91 | 1.57 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDDYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------- | |||||||||||||||||||
| 7 | 4djh | 0.13 | 0.14 | 0.92 | 1.71 | Download | --PAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIE--CSLQFPDDWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF-------------- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.19 | 0.91 | 2.23 | Download | TDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR---KKSLQSTVDYYLGSLALSDLLILLLVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIP--MLFTMGLQNLSGDGTHPGGVCTPITATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ---------PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL--------------- | |||||||||||||||||||
| 9 | 4djhA | 0.13 | 0.18 | 0.89 | 1.50 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF-------------P | |||||||||||||||||||
| 10 | 1l9hA | 0.12 | 0.19 | 0.94 | 1.56 | Download | AEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQ----HKKLTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCPMSNFRF------GENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSYYTPHEETNNESFVIYMFVVHFIIPLIVIFFCYGQLVFTV----------KEAAAATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDSTTVSK | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
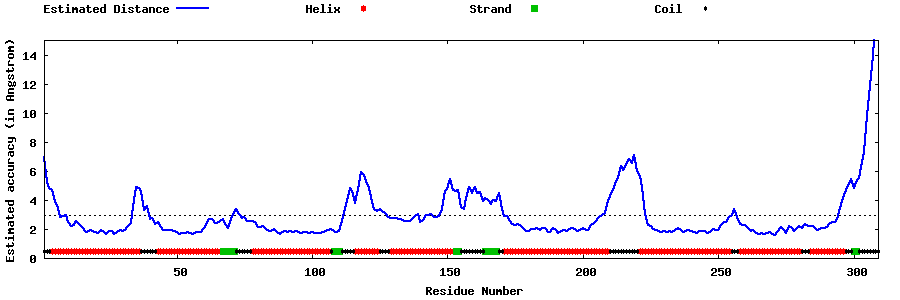 | |||
|
|
|||
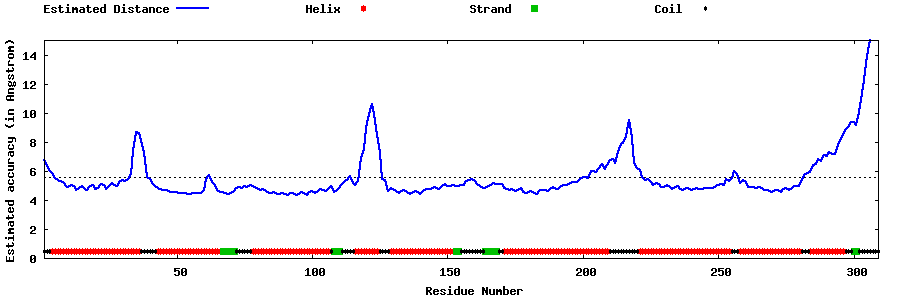 | |||
|
| |||
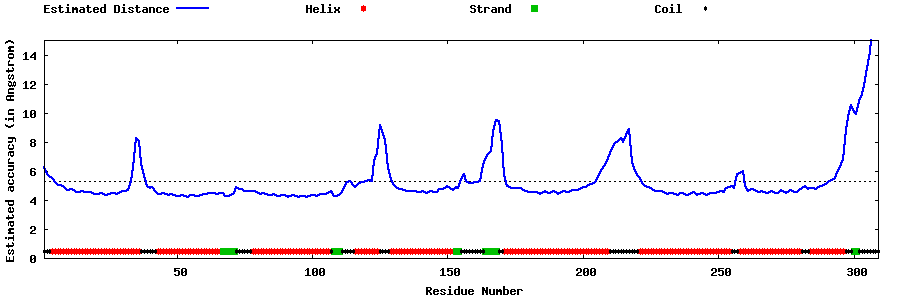 | |||
|
| |||
 | |||
|
| |||
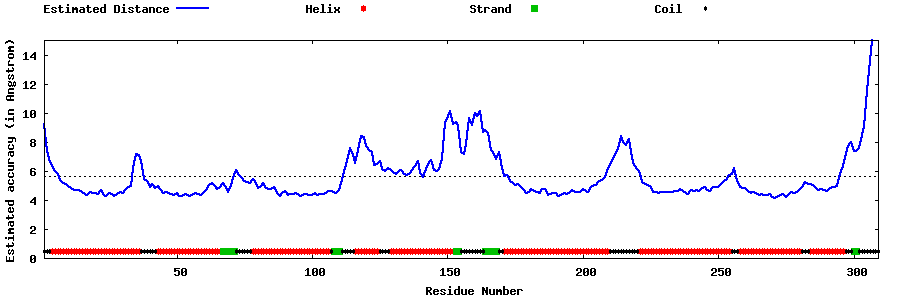 | |||
|
| |||