

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MITFLPIIFSILIVVIFVIGNFANGFIALVNSIEWVKRQKISFVDQILTALAVSRVGLLWVLLLHWYATQLNPAFYSVEVRITAYNVWAVTNHFSSWLATSLSMFYLLRIANFSNLIFLRIKRRVKSVVLVILLGPLLFLVCHLFVINMDETVWTKEYEGNVTWKIKLRSAMYHSNMTLTMLANFVPLTLTLISFLLLICSLCKHLKKMQLHGKGSQDPSTKVHIKALQTVTSFLLLCAIYFLSMIISVCNFGRLEKQPVFMFCQAIIFSYPSTHPFILILGNKKLKQIFLSVLRHVRYWVKDRSLRLHRFTRGALCVF | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCSSCC | |
| 9758999999999999999999999999999999983797876759999999999999999997172486342110517899999899998199999999999997078607996799999997217299999999999999999985422202467888421676501358899999999999999999999999999999999999737799899998089999999999999999999999999999875026659999999999982788789895187199999999999645033899998873687650129 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MITFLPIIFSILIVVIFVIGNFANGFIALVNSIEWVKRQKISFVDQILTALAVSRVGLLWVLLLHWYATQLNPAFYSVEVRITAYNVWAVTNHFSSWLATSLSMFYLLRIANFSNLIFLRIKRRVKSVVLVILLGPLLFLVCHLFVINMDETVWTKEYEGNVTWKIKLRSAMYHSNMTLTMLANFVPLTLTLISFLLLICSLCKHLKKMQLHGKGSQDPSTKVHIKALQTVTSFLLLCAIYFLSMIISVCNFGRLEKQPVFMFCQAIIFSYPSTHPFILILGNKKLKQIFLSVLRHVRYWVKDRSLRLHRFTRGALCVF | |
| 7332232211321331233133102200210111004434032011000000111000000133300000000202333000000000110122000000000100000002231100000133043100110121223333220213033313264342111131313332201111133333331331133133102100220232024334434113040022001001202211331231011010122334430010101121021010101010131430040023002101010464514133333300003 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCSSSSSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCSSCC MITFLPIIFSILIVVIFVIGNFANGFIALVNSIEWVKRQKISFVDQILTALAVSRVGLLWVLLLHWYATQLNPAFYSVEVRITAYNVWAVTNHFSSWLATSLSMFYLLRIANFSNLIFLRIKRRVKSVVLVILLGPLLFLVCHLFVINMDETVWTKEYEGNVTWKIKLRSAMYHSNMTLTMLANFVPLTLTLISFLLLICSLCKHLKKMQLHGKGSQDPSTKVHIKALQTVTSFLLLCAIYFLSMIISVCNFGRLEKQPVFMFCQAIIFSYPSTHPFILILGNKKLKQIFLSVLRHVRYWVKDRSLRLHRFTRGALCVF | |||||||||||||||||||||||||
| 1 | 4djhA | 0.12 | 0.16 | 0.87 | 1.37 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSDR-----------NLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------- | |||||||||||||||||||
| 2 | 5tjvA | 0.11 | 0.19 | 0.85 | 2.88 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHS--RSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSDIFPHID------------ETYLMFWIGVTSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------------------- | |||||||||||||||||||
| 3 | 4djhA | 0.12 | 0.16 | 0.86 | 2.19 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY--TKMK-TATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKAL----DFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL-------------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------- | |||||||||||||||||||
| 4 | 4ib4 | 0.14 | 0.22 | 0.89 | 1.55 | Download | EQGNKLHWAALLILMVIIPTIGGNTLVILAVSL---EKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCAWLFLDVLFSTASIWHLCAISVDRYIAIK---KPIQAN-QYNSRATAFIKITVVWLIIGIAIPVPIGIETNPNNTCVLTKE-----RFGDFML--FGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-------------------- | |||||||||||||||||||
| 5 | 3odu | 0.15 | 0.26 | 0.93 | 1.27 | Download | NANFNKIFLPTIYSIIFLTGIVGNGLVILVMGY---QKKLRSMTDKYRLHLSVADLLFVIT-LPFWAVDAVANWYFGNFLCKAVHVIYTVNLYSSVWILAFISLDRYLAIVHATNSQRP----RKLLAEKVVYVGVWIPALLLTIPDFIFANVSEADDRYICDRFYPNDLWVVVFQFQHIMVGLILPGIVILSCYCIIISKLSHSGSNIFEMLRDAYGSKGHQKRKALKTTVILILAFFACWLPYYIGISIDFILLVHKWISITEALAFFHCCLNPILYAFLGAKFKTSAQHALTSGRPLEVLFQ-------------- | |||||||||||||||||||
| 6 | 4djhA | 0.12 | 0.16 | 0.87 | 1.51 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA----LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------- | |||||||||||||||||||
| 7 | 4djh | 0.12 | 0.16 | 0.89 | 1.70 | Download | --PAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDV--DVIECSLQFPDDDWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGS---AALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF------------------------ | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.19 | 0.88 | 2.38 | Download | TDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR---KKSLQSTVDYYLGSLALSDLLILLLVELYNFIWVHHPWAFGDAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMG-LQNLSGDGTHPGGLVCTPITATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQ---------PGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------------------------- | |||||||||||||||||||
| 9 | 4djhA | 0.12 | 0.16 | 0.86 | 1.49 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQFPWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLL------------DRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP----------------------- | |||||||||||||||||||
| 10 | 6b73A | 0.12 | 0.15 | 0.83 | 1.34 | Download | ISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKAPLKAKIINICIWLLSSSVGISAIVLGGTKVRDVISLQFPDDDYSW------WDLFMKICVFIFAFVIPVLIIIVCYTLMILRLK-------SVRSREKD---RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGTSLSSYYFCIALGYTNSSLNPILYAFLDENFKR------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
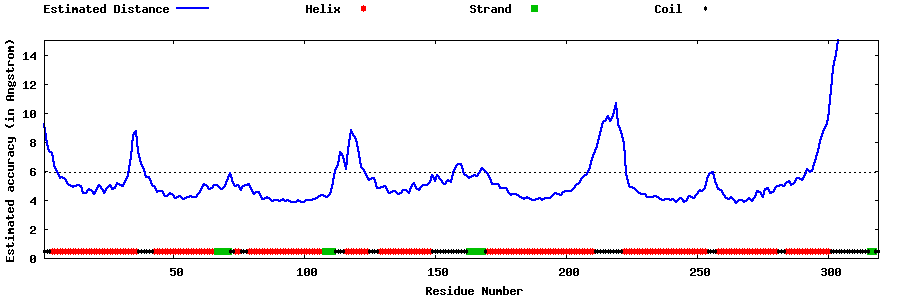
| Generated 3D models | Estimated local accuracy of models | ||
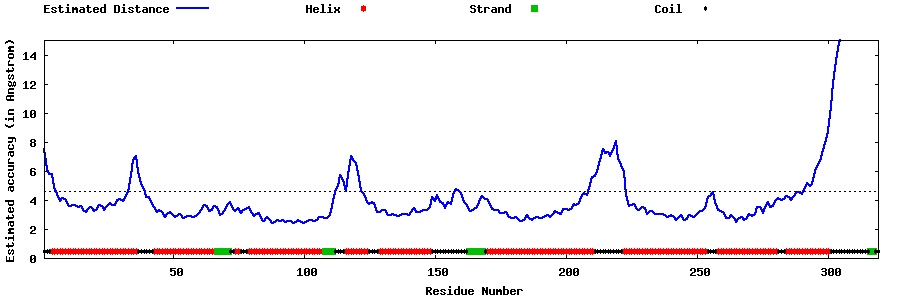 | |||
|
|
|||
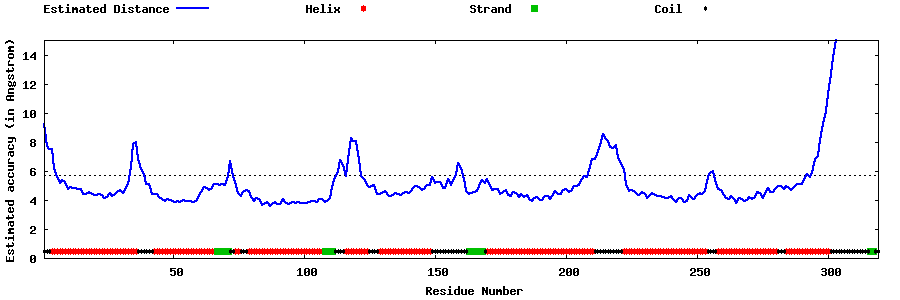 | |||
|
| |||
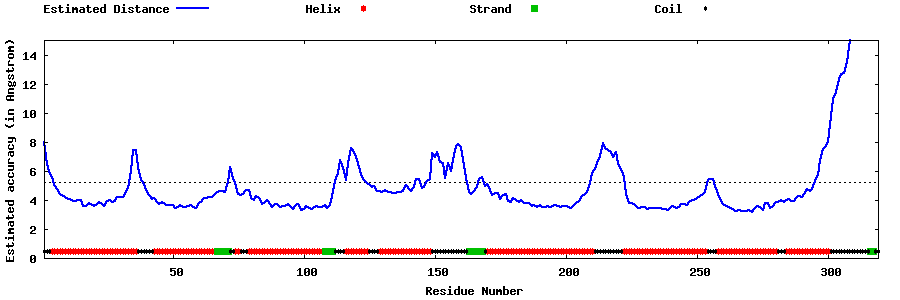 | |||
|
| |||
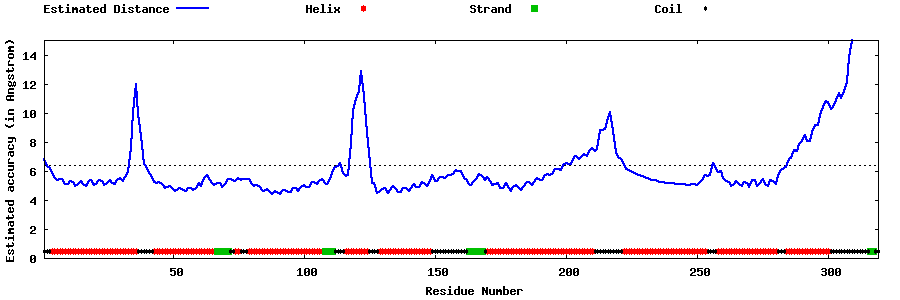 | |||
|
| |||
 | |||
|
| |||