

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSKRSWWAGSRKPPREMLKLSGSDSSQSMNGLEVAPPGLITNFSLATAEQCGQETPLENMLFASFYLLDFILALVGNTLALWLFIRDHKSGTPANVFLMHLAVADLSCVLVLPTRLVYHFSGNHWPFGEIACRLTGFLFYLNMYASIYFLTCISADRFLAIVHPVKSLKLRRPLYAHLACAFLWVVVAVAMAPLLVSPQTVQTNHTVVCLQLYREKASHHALVSLAVAFTFPFITTVTCYLLIIRSLRQGLRVEKRLKTKAVRMIAIVLAIFLVCFVPYHVNRSVYVLHYRSHGASCATQRILALANRITSCLTSLNGALDPIMYFFVAEKFRHALCNLLCGKRLKGPPPSFEGKTNESSLSAKSEL | |
| CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCSSCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 9866678999899521245888887777777877897657778888777788750168987999999999999999999889766236778699999999999999999973999999966899887867899999999999999999999999983799970401023655046888899999999999999999600362699158607396656999999999989999999999999999998025466653224499999999999999866899999999999746789827999999999999999999998889988820888999999999762468998666888888888766889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | |
| | | | | | | | | | | | | | | | | | | | |
| MSKRSWWAGSRKPPREMLKLSGSDSSQSMNGLEVAPPGLITNFSLATAEQCGQETPLENMLFASFYLLDFILALVGNTLALWLFIRDHKSGTPANVFLMHLAVADLSCVLVLPTRLVYHFSGNHWPFGEIACRLTGFLFYLNMYASIYFLTCISADRFLAIVHPVKSLKLRRPLYAHLACAFLWVVVAVAMAPLLVSPQTVQTNHTVVCLQLYREKASHHALVSLAVAFTFPFITTVTCYLLIIRSLRQGLRVEKRLKTKAVRMIAIVLAIFLVCFVPYHVNRSVYVLHYRSHGASCATQRILALANRITSCLTSLNGALDPIMYFFVAEKFRHALCNLLCGKRLKGPPPSFEGKTNESSLSAKSEL | |
| 7544434426443344134043343343133343334332331323444414344402100002101200230231122001000122343210100000001000000000101000003532010021011001101130110001000000310000001002023133221000000000100000000100013036573221010213642110101100212331220002000100110242354654411000000000000010013210100001002313334414133002101100100003000200100000045014101400232314444443444455444345367 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCSSCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCCC MSKRSWWAGSRKPPREMLKLSGSDSSQSMNGLEVAPPGLITNFSLATAEQCGQETPLENMLFASFYLLDFILALVGNTLALWLFIRDHKSGTPANVFLMHLAVADLSCVLVLPTRLVYHFSGNHWPFGEIACRLTGFLFYLNMYASIYFLTCISADRFLAIVHPVKSLKLRRPLYAHLACAFLWVVVAVAMAPLLVSPQTVQTNHTVVCLQLYREKASHHALVSLAVAFTFPFITTVTCYLLIIRSLRQGLRVEKRLKTKAVRMIAIVLAIFLVCFVPYHVNRSVYVLHYRSHGASCATQRILALANRITSCLTSLNGALDPIMYFFVAEKFRHALCNLLCGKRLKGPPPSFEGKTNESSLSAKSEL | |||||||||||||||||||||||||
| 1 | 4mbsA | 0.24 | 0.20 | 0.79 | 3.22 | Download | -----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPYWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLR-MKEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 2 | 4mbsA | 0.24 | 0.20 | 0.79 | 4.24 | Download | ------------------------------------------------CQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMEVEEEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFF--------------------------- | |||||||||||||||||||
| 3 | 4mbsA | 0.24 | 0.20 | 0.79 | 3.81 | Download | -----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAAQ--WDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKE-EEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 4 | 4djh | 0.28 | 0.25 | 0.75 | 1.55 | Download | ------------------------------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTTTMPFQSTVYLM-NSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKRDVDVIECSLQFPDDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA-----------ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------------- | |||||||||||||||||||
| 5 | 5t1a | 0.26 | 0.24 | 0.77 | 1.18 | Download | -----------------------------------------------------VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE-DSVYVCGPYFPRGNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF--------------------------- | |||||||||||||||||||
| 6 | 4mbsA | 0.24 | 0.20 | 0.79 | 3.32 | Download | -------------------------------------------------QKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKEEE-KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 7 | 5t1a | 0.26 | 0.24 | 0.77 | 1.70 | Download | -------------------------------------------------------QIGAQLLPPLYSLVFIFGFVGNMLVVLILINCKKLKCLTDIYLLNLAISDLLFLITLPLWAHSAA--NEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFALKARTVTFGVVTSVITWLVAVFASVPGIIFTKQK-EDSVYVCGPYFPRGNNFHTIMRNILGLVLPLLIMVICYSGISRASKSRINIPPSREKKAVRVIFTIMIVYFLFWTPYNIVILLNTFQEFFGLSNCESTSQLDQATQVTETLGMTHCCINPIIYAFVGEKFRRYLSVFF--------------------------- | |||||||||||||||||||
| 8 | 4mbsA | 0.24 | 0.20 | 0.78 | 4.94 | Download | -----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAAA--QWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRE-----KKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 9 | 4mbsA | 0.24 | 0.20 | 0.79 | 3.46 | Download | -----------------------------------------------PCQKINVKQIAARLLPPLYSLVFIFGFVGNMLVILILINYKRLKSMTDIYLLNLAISDLFFLLTVPFWAHYAA--AQWDFGNTMCQLLTGLYFIGFFSGIFFIILLTIDRYLAVVHAVFALKARTVTFGVVTSVITWVVAVFASLPNIIFTRSQKEGLHYTCSSHFPFWKNFQTLKIVILGLVLPLLVMVICYSGILKTLLRMKE-EEKKRHRDVRLIFTIMIVYFLFWAPYNIVLLLNTFQEFFGLNNCSSSNRLDQAMQVTETLGMTHCCINPIIYAFVGEEFRNYLLVFFQ-------------------------- | |||||||||||||||||||
| 10 | 5c1mA | 0.26 | 0.21 | 0.78 | 3.96 | Download | -------------------------------------------GSHSLPQTGSPSMVTAITIMALYSIVCVVGLFGNFLVMYVIVRYTKMKTATNIYIFNLALADALATSTLPFQSVNYLMG-TWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFRTPRNAKIVNVCNWILSSAIGLPVMFMATTKYRQGSIDCTLTFSYWENLLKICVFIFAFIMPVLIITVCYGLMILRLKSVRMLSGSKLRRITRMVLVVVAVFIVCWTPIHIYVIIKALITIPET------TFQTVSWHFCIALGYTNSCLNPVLYAFLDENFKRCF------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
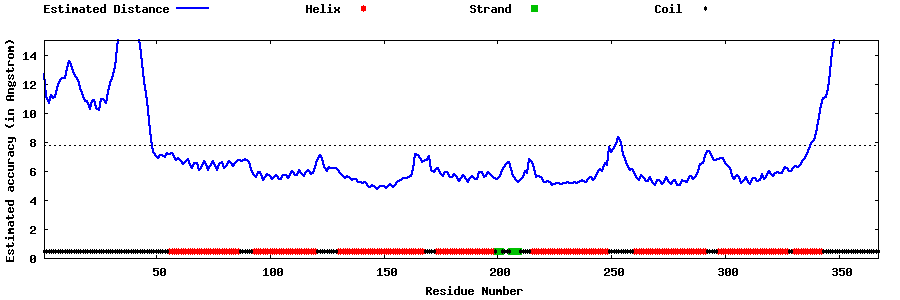
| Generated 3D models | Estimated local accuracy of models | ||
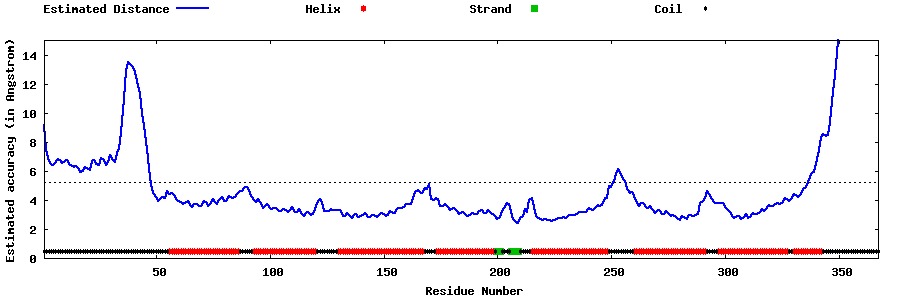 | |||
|
|
|||
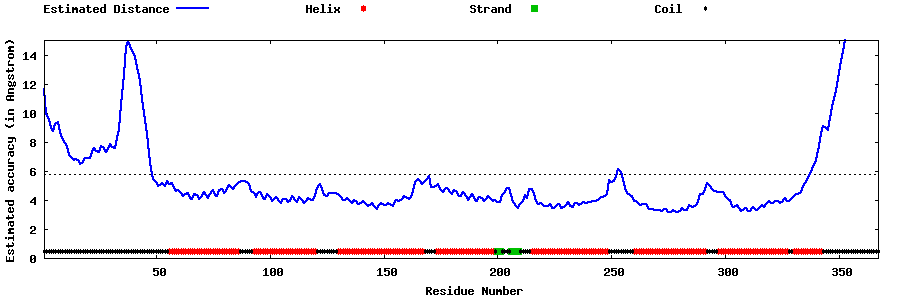 | |||
|
| |||
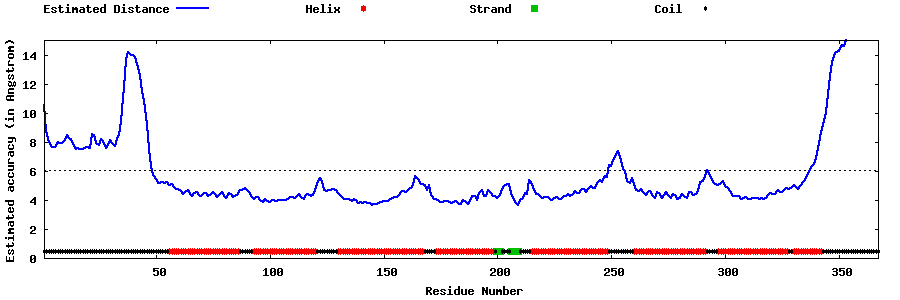 | |||
|
| |||
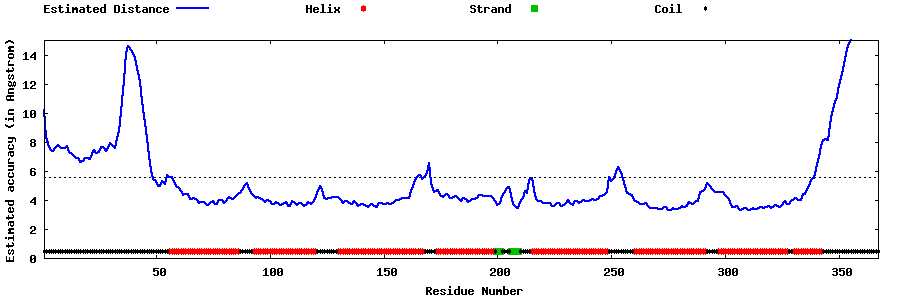 | |||
|
| |||
 | |||
|
| |||