

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MGDVNQSVASDFILVGLFSHSGSRQLLFSLVAVMFVIGLLGNTVLLFLIRVDSRLHTPMYFLLSQLSLFDIGCPMVTIPKMASDFLRGEGATSYGGGAAQIFFLTLMGVAEGVLLVLMSYDRYVAVCQPLQYPVLMRRQVCLLMMGSSWVVGVLNASIQTSITLHFPYCASRIVDHFFCEVPALLKLSCADTCAYEMALSTSGVLILMLPLSLIATSYGHVLQAVLSMRSEEARHKAVTTCSSHITVVGLFYGAAVFMYMVPCAYHSPQQDNVVSLFYSLVTPTLNPLIYSLRNPEVWMALVKVLSRAGLRQMC | |
| CCCCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCHHHCC | |
| 99988730233578779998125899999999999999998999999998677745639999888999987545005899999835799578688899999999999999999999998650776264446883258869999999999999999999999992068899995888524858888897037519999999999999999899999999999999824876132630764568789989999986886150789999976692888337206544024533156499999999998431231069 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MGDVNQSVASDFILVGLFSHSGSRQLLFSLVAVMFVIGLLGNTVLLFLIRVDSRLHTPMYFLLSQLSLFDIGCPMVTIPKMASDFLRGEGATSYGGGAAQIFFLTLMGVAEGVLLVLMSYDRYVAVCQPLQYPVLMRRQVCLLMMGSSWVVGVLNASIQTSITLHFPYCASRIVDHFFCEVPALLKLSCADTCAYEMALSTSGVLILMLPLSLIATSYGHVLQAVLSMRSEEARHKAVTTCSSHITVVGLFYGAAVFMYMVPCAYHSPQQDNVVSLFYSLVTPTLNPLIYSLRNPEVWMALVKVLSRAGLRQMC | |
| 86442302202000000043241110001203332331333333002001001300000011021000000111002002000000156310101002201120131132012000300300000002102000100330000001200310031030001100001103424010000032100200020033002102331233133123103201220020011031450231011000000000001320000000103243255241000011023203300300001042023002200343314422 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCHHHCC MGDVNQSVASDFILVGLFSHSGSRQLLFSLVAVMFVIGLLGNTVLLFLIRVDSRLHTPMYFLLSQLSLFDIGCPMVTIPKMASDFLRGEGATSYGGGAAQIFFLTLMGVAEGVLLVLMSYDRYVAVCQPLQYPVLMRRQVCLLMMGSSWVVGVLNASIQTSITLHFPYCASRIVDHFFCEVPALLKLSCADTCAYEMALSTSGVLILMLPLSLIATSYGHVLQAVLSMRSEEARHKAVTTCSSHITVVGLFYGAAVFMYMVPCAYHSPQQDNVVSLFYSLVTPTLNPLIYSLRNPEVWMALVKVLSRAGLRQMC | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.22 | 0.89 | 3.55 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.17 | 0.19 | 0.89 | 2.27 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKLQSC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM---------- | |||||||||||||||||||
| 3 | 4iaqA | 0.16 | 0.20 | 0.85 | 2.16 | Download | ------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAFY---FPTLLLIALYGRIYVEARSQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.18 | 0.89 | 1.54 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------- | |||||||||||||||||||
| 5 | 4yay | 0.14 | 0.21 | 0.90 | 1.24 | Download | KTT-RNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.22 | 0.88 | 3.78 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.20 | 0.83 | 1.73 | Download | -------------------SLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASECVVN----------T-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTL---GIILGAFIVCWLPFFLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 8 | 2z73A | 0.17 | 0.21 | 0.94 | 2.56 | Download | --ETNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCNDYISR-------------DSTTRSNILCMFILG---FFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCC-Q | |||||||||||||||||||
| 9 | 3emlA | 0.18 | 0.22 | 0.89 | 4.49 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKS-LAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.20 | 0.92 | 5.49 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQE | |||||||||||||||||||
| ||||||||||||||||||||||||||
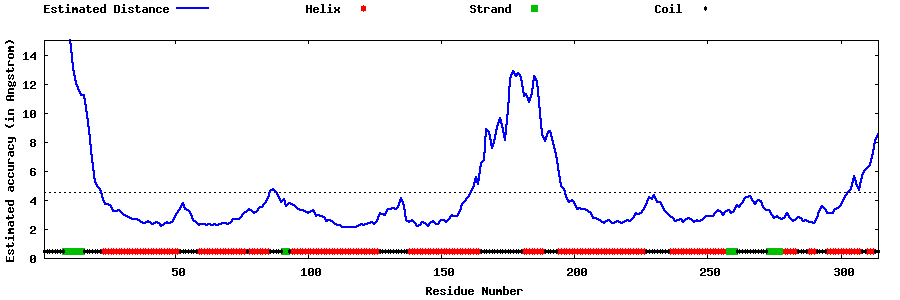
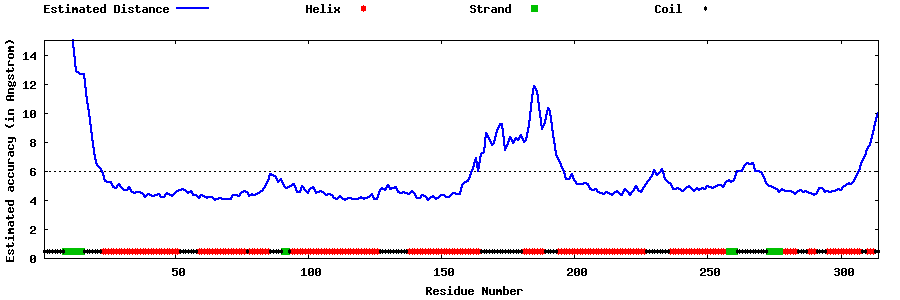
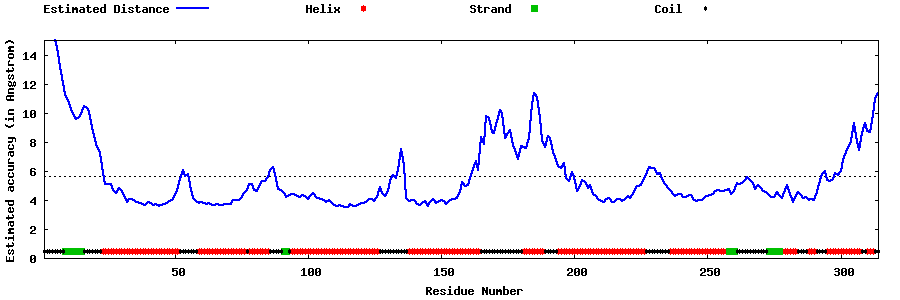
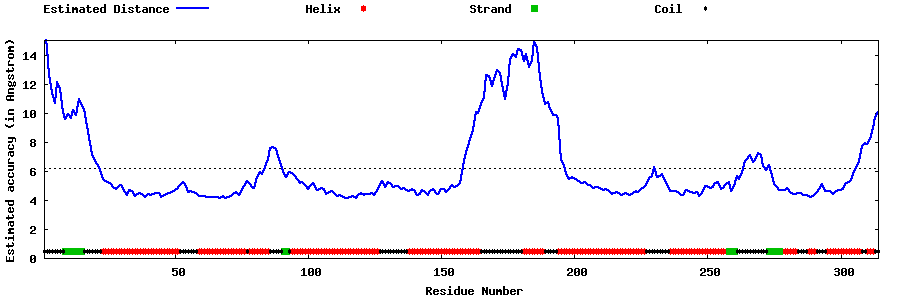
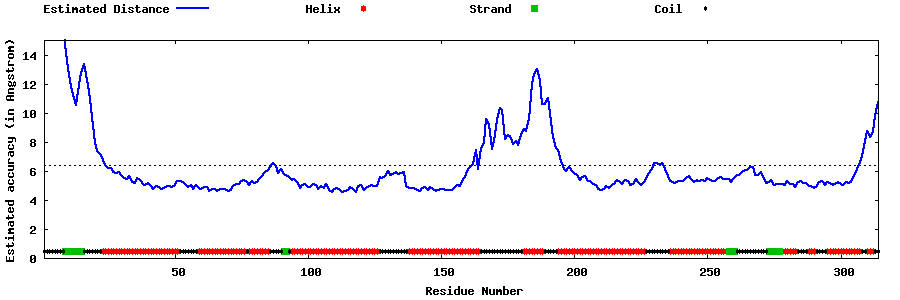
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||