

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MEPRNQTSASQFILLGLSEKPEQETLLFSLFFCMYLVMVVGNLLIILAISIDSHLHTPMYFFLANLSLVDFCLATNTIPKMLVSLQTGSKAISYPCCLIQMYFFHFFGIVDSVIIAMMAYDRFVAICHPLHYAKIMSLRLCRLLVGALWAFSCFISLTHILLMARLVFCGSHEVPHYFCDLTPILRLSCTDTSVNRIFILIVAGMVIATPFVCILASYARILVAIMKVPSAGGRKKAFSTCSSHLSVVALFYGTTIGVYLCPSSVLTTVKEKASAVMYTAVTPMLNPFIYSLRNRDLKGALRKLVNRKITSSS | |
| CCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSCCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHCCCCCCCC | |
| 9998885075352116998820568999999999999999899999999628875552999988799986523404789988998448974758999999999999999899999999875144126754377423772899999999999999999999999741589999568936785998766164426989999999999999869999999999999841136878867520570778999999857032366078987889888289984016462434564246618899999999704457789 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MEPRNQTSASQFILLGLSEKPEQETLLFSLFFCMYLVMVVGNLLIILAISIDSHLHTPMYFFLANLSLVDFCLATNTIPKMLVSLQTGSKAISYPCCLIQMYFFHFFGIVDSVIIAMMAYDRFVAICHPLHYAKIMSLRLCRLLVGALWAFSCFISLTHILLMARLVFCGSHEVPHYFCDLTPILRLSCTDTSVNRIFILIVAGMVIATPFVCILASYARILVAIMKVPSAGGRKKAFSTCSSHLSVVALFYGTTIGVYLCPSSVLTTVKEKASAVMYTAVTPMLNPFIYSLRNRDLKGALRKLVNRKITSSS | |
| 8567231200200001015416212000010030122023312000000100230000000000010110111200310300100114334030200000110002002201000000021000000000200000002000000120020002002100000200010372400000012320030001102101010122133303323300320101000000102146232100000000000000121002000000121314432200000001312330010000203102300230033333348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSCCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHCCCCCCCC MEPRNQTSASQFILLGLSEKPEQETLLFSLFFCMYLVMVVGNLLIILAISIDSHLHTPMYFFLANLSLVDFCLATNTIPKMLVSLQTGSKAISYPCCLIQMYFFHFFGIVDSVIIAMMAYDRFVAICHPLHYAKIMSLRLCRLLVGALWAFSCFISLTHILLMARLVFCGSHEVPHYFCDLTPILRLSCTDTSVNRIFILIVAGMVIATPFVCILASYARILVAIMKVPSAGGRKKAFSTCSSHLSVVALFYGTTIGVYLCPSSVLTTVKEKASAVMYTAVTPMLNPFIYSLRNRDLKGALRKLVNRKITSSS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.20 | 0.89 | 3.67 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.21 | 0.90 | 2.16 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 3 | 5tjvA | 0.20 | 0.22 | 0.89 | 2.02 | Download | ---ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLCSDIF-PHID------------------ETYLMFWIGVTSVLLLFIVYAYMYILWKAMSFSDQRMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCSTVNPIIYALRSKDLRHAFRSMF-------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.23 | 0.89 | 1.55 | Download | -----------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIE------TNPNNITCVLTK--------E-RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADSNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDNLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR--- | |||||||||||||||||||
| 5 | 4yay | 0.18 | 0.22 | 0.90 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNV-FF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL-------- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.20 | 0.88 | 3.87 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.23 | 0.83 | 1.73 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQAS----------ECVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADLRERKATKTL---GIILGAFIVCWLPFFLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK----- | |||||||||||||||||||
| 8 | 2z73A | 0.20 | 0.21 | 0.96 | 3.01 | Download | -ETYNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFG------WGAYTLEGVLCD-------YISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQ | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.20 | 0.89 | 5.16 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ- | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.20 | 0.92 | 5.85 | Download | -----------------S------SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
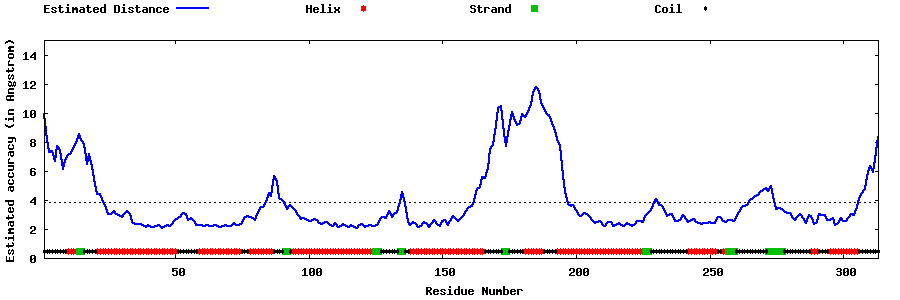 | |||
|
|
|||
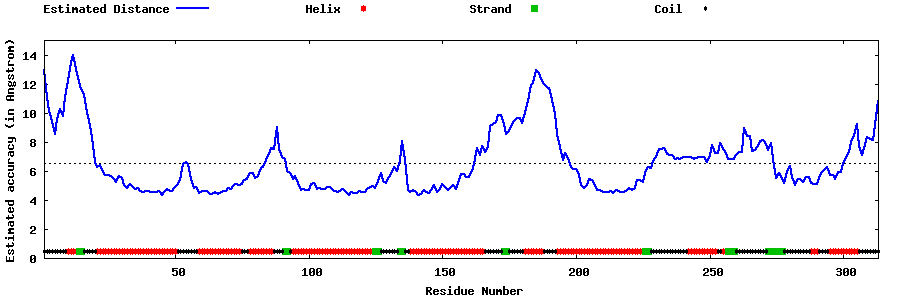 | |||
|
| |||
 | |||
|
| |||
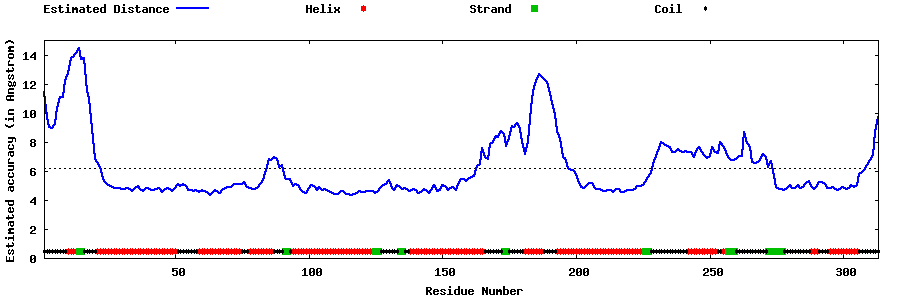 | |||
|
| |||
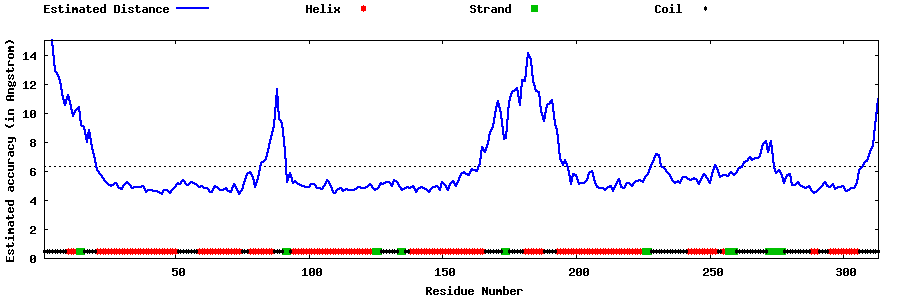 | |||
|
| |||