

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MEFVFLAYPSCPELHILSFLGVSLVYGLIITGNILIVVSIHTETCLCTSMYYFLGSLSGIEICYTAVVVPHILANTLQSEKTITLLGCATQMAFFIALGSADCFLLAAMAYDRYVAICHPLQYPLLMTLTLCVHLVVASVISGLFLSLQLVAFIFSLPFCQAQGIEHFFCDVPPVMHVVCAQSHIHEQSVLVAAILAIAVPFFLITTSYTFIVAALLKIHSAAGRHRAFSTCSSHLTVVLLQYGCCAFMYLCPSSSYNPKQDRFISLVYTLGTPLLNPLIYALRNSEMKGAVGRVLTRNCLSQNS | |
| CSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCC | |
| 93899628998627999999999999999998899978887288756748889878999988887206899999856799778489999999999999999999999998650564065444885148879999999999999999999999980548899796788624838878885358719999999999999999799999999999999842866154641542568789979998844640680789999887782899866336742023543046499999999998615566679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MEFVFLAYPSCPELHILSFLGVSLVYGLIITGNILIVVSIHTETCLCTSMYYFLGSLSGIEICYTAVVVPHILANTLQSEKTITLLGCATQMAFFIALGSADCFLLAAMAYDRYVAICHPLQYPLLMTLTLCVHLVVASVISGLFLSLQLVAFIFSLPFCQAQGIEHFFCDVPPVMHVVCAQSHIHEQSVLVAAILAIAVPFFLITTSYTFIVAALLKIHSAAGRHRAFSTCSSHLTVVLLQYGCCAFMYLCPSSSYNPKQDRFISLVYTLGTPLLNPLIYALRNSEMKGAVGRVLTRNCLSQNS | |
| 52000000043340101001302331331333332001002003300100011020001102201001203000101166330102002201100010021001000100100000002102000200330000000000110031031101100002003423010000032000200021031002211331233133123103302320000001031463232000000001000101120000000103163346341000031033203300300001142014002200233313468 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCCCCC MEFVFLAYPSCPELHILSFLGVSLVYGLIITGNILIVVSIHTETCLCTSMYYFLGSLSGIEICYTAVVVPHILANTLQSEKTITLLGCATQMAFFIALGSADCFLLAAMAYDRYVAICHPLQYPLLMTLTLCVHLVVASVISGLFLSLQLVAFIFSLPFCQAQGIEHFFCDVPPVMHVVCAQSHIHEQSVLVAAILAIAVPFFLITTSYTFIVAALLKIHSAAGRHRAFSTCSSHLTVVLLQYGCCAFMYLCPSSSYNPKQDRFISLVYTLGTPLLNPLIYALRNSEMKGAVGRVLTRNCLSQNS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.23 | 0.24 | 0.91 | 3.72 | Download | -----------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINC-FTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.23 | 0.88 | 2.16 | Download | ----------NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEKL---------QSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM---------- | |||||||||||||||||||
| 3 | 4iaqA | 0.16 | 0.20 | 0.86 | 2.17 | Download | ---YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAF---YFPTLLLIALYGRIYVEARSRIIQKYAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.25 | 0.92 | 1.54 | Download | ----------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------- | |||||||||||||||||||
| 5 | 4yay | 0.20 | 0.27 | 0.90 | 1.22 | Download | QKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------- | |||||||||||||||||||
| 6 | 3emlA | 0.23 | 0.24 | 0.91 | 3.87 | Download | ------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.23 | 0.86 | 1.71 | Download | ------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPI--HL-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 8 | 2z73A | 0.13 | 0.22 | 0.93 | 2.34 | Download | REF-----DQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGTLEGVLCNDYI----------------SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQF | |||||||||||||||||||
| 9 | 3emlA | 0.22 | 0.24 | 0.91 | 4.99 | Download | -----------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.23 | 0.95 | 6.03 | Download | --------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQE | |||||||||||||||||||
| ||||||||||||||||||||||||||
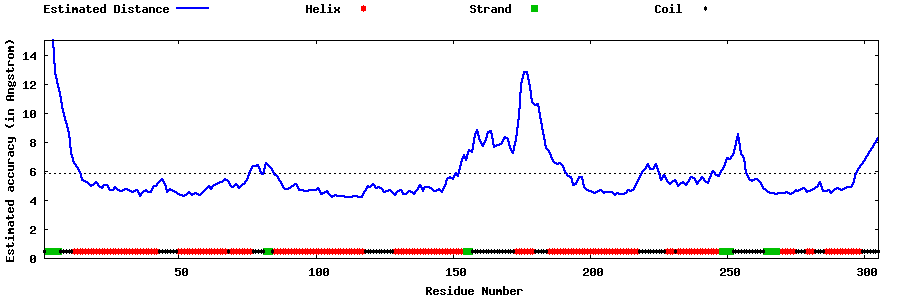
| Generated 3D models | Estimated local accuracy of models | ||
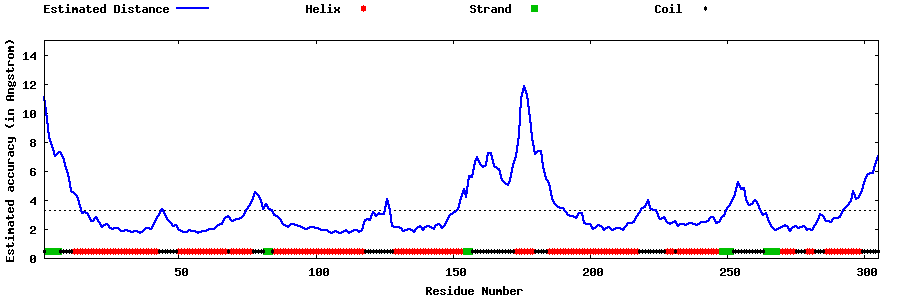 | |||
|
|
|||
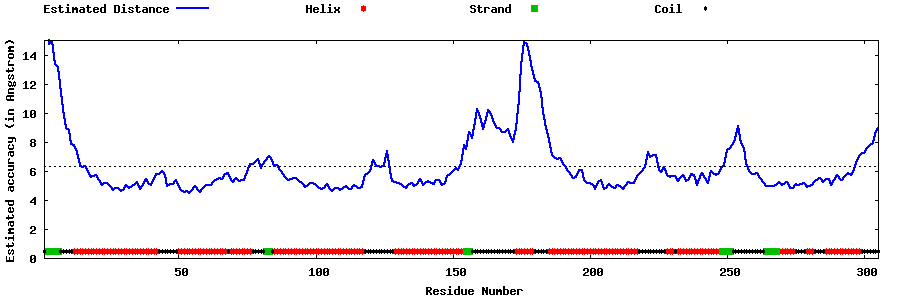 | |||
|
| |||
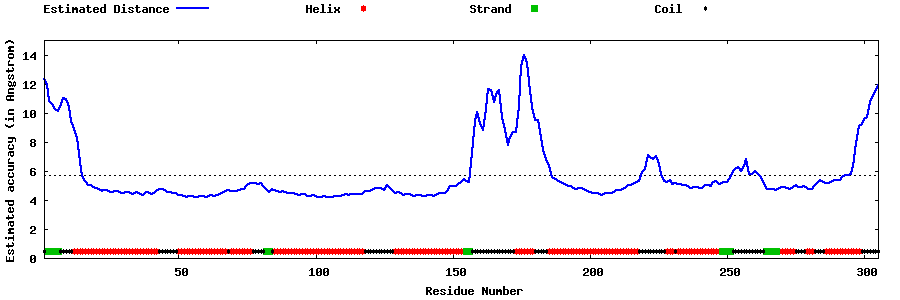 | |||
|
| |||
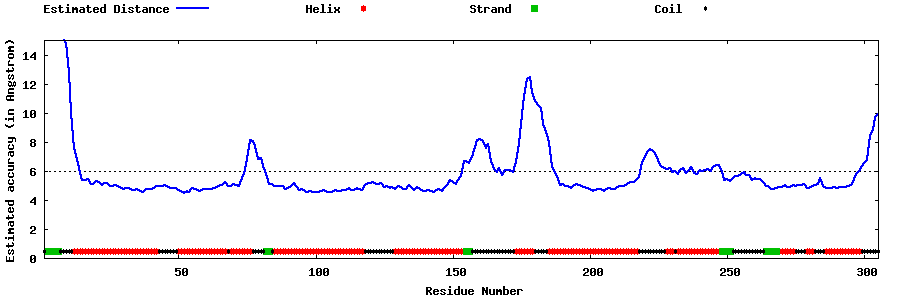 | |||
|
| |||
 | |||
|
| |||