

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MEPRKNVTDFVLLGFTQNPKEQKVLFVMFLLFYILTMVGNLLIVVTVTVSETLGSPMSFFLAGLTFIDIIYSSSISPRLISDLFFGNNSISFQSFMAQLFIEHLFGGSEVFLLLVMAYDRYVAICKPLHYLVIMRQWVCVLLLVVSWVGGFLQSVFQLSIIYGLPFCGPNVIDHFFCDMYPLLKLACTDTHVIGLLVVANGGLSCTIAFLLLLISYGVILHSLKKLSQKGRQKAHSTCSSHITVVVFFFVPCIFMCARPARTFSIDKSVSVFYTVITPMLNPLIYTLRNSEMTSAMKKL | |
| CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCHHHHHHHHHHHHHCCCHHHCCCCHHHHHHHHCC | |
| 99997210568806999812589999999999999998788745523207999997999987499875533234719999987439967838999999999999999999999999965178746210262126775999999999998999999999992458998988367764383888889402405322004446449999999999999999999998856421209999998799933256206435884889999750000677763364010374335629999998529 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MEPRKNVTDFVLLGFTQNPKEQKVLFVMFLLFYILTMVGNLLIVVTVTVSETLGSPMSFFLAGLTFIDIIYSSSISPRLISDLFFGNNSISFQSFMAQLFIEHLFGGSEVFLLLVMAYDRYVAICKPLHYLVIMRQWVCVLLLVVSWVGGFLQSVFQLSIIYGLPFCGPNVIDHFFCDMYPLLKLACTDTHVIGLLVVANGGLSCTIAFLLLLISYGVILHSLKKLSQKGRQKAHSTCSSHITVVVFFFVPCIFMCARPARTFSIDKSVSVFYTVITPMLNPLIYTLRNSEMTSAMKKL | |
| 87442200000000004245001000131333233233323300000200430400001002300120011000000200000036321000500000001103311310200020030000000221101000233100000120131023013001310230300010220001002210030000202300030023013202310331331121002001321461222011003002100202321010000003332330001003102310331230000205402301455 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCHHHCHHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCHHHHHHHHHHHHHCCCHHHCCCCHHHHHHHHCC MEPRKNVTDFVLLGFTQNPKEQKVLFVMFLLFYILTMVGNLLIVVTVTVSETLGSPMSFFLAGLTFIDIIYSSSISPRLISDLFFGNNSISFQSFMAQLFIEHLFGGSEVFLLLVMAYDRYVAICKPLHYLVIMRQWVCVLLLVVSWVGGFLQSVFQLSIIYGLPFCGPNVIDHFFCDMYPLLKLACTDTHVIGLLVVANGGLSCTIAFLLLLISYGVILHSLKKLSQKGRQKAHSTCSSHITVVVFFFVPCIFMCARPARTFSIDKSVSVFYTVITPMLNPLIYTLRNSEMTSAMKKL | |||||||||||||||||||||||||
| 1 | 5tgzA | 0.20 | 0.24 | 0.93 | 3.35 | Download | GENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG-------NCEKLQSVCSDIFPH----------IDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.24 | 0.92 | 2.26 | Download | -ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM | |||||||||||||||||||
| 3 | 5tgzA | 0.18 | 0.24 | 0.93 | 2.21 | Download | -GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSDIFPHID-------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPRMDIELAKTLVLILVVLIICWGPLLAIMVGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM | |||||||||||||||||||
| 4 | 4djh | 0.13 | 0.21 | 0.92 | 1.55 | Download | -----------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSLSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDF | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.22 | 0.93 | 1.22 | Download | KTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVGRVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQL | |||||||||||||||||||
| 6 | 5tgzA | 0.21 | 0.24 | 0.93 | 3.70 | Download | -ENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAV--------LPLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQAIELAKTLVLILVVLIICWGPLLAIMVYDVFGIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.25 | 0.62 | 1.72 | Download | -------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASE----------CVVNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSR------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.15 | 0.19 | 0.91 | 2.70 | Download | ---NSD------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGVCTPI--------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQMLTNALVYVSAAINPILYNLVSANFRQVFLST | |||||||||||||||||||
| 9 | 5tgzA | 0.21 | 0.24 | 0.92 | 4.84 | Download | GENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.21 | 0.92 | 5.41 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKI | |||||||||||||||||||
| ||||||||||||||||||||||||||
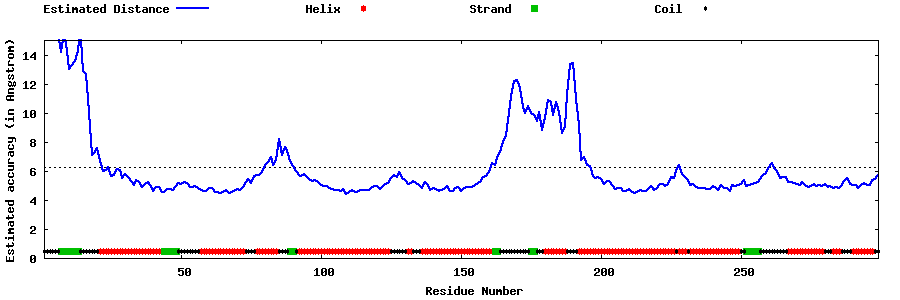
| Generated 3D models | Estimated local accuracy of models | ||
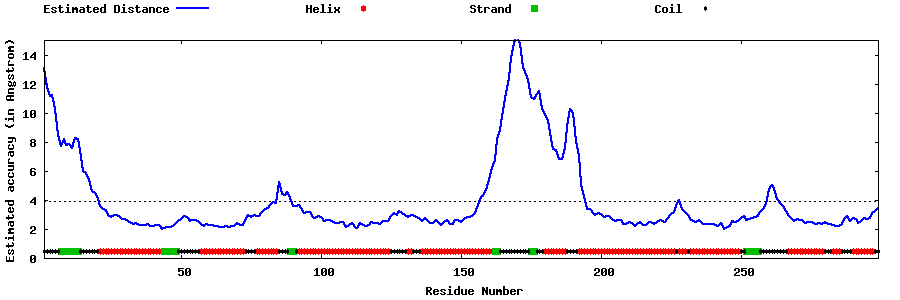 | |||
|
|
|||
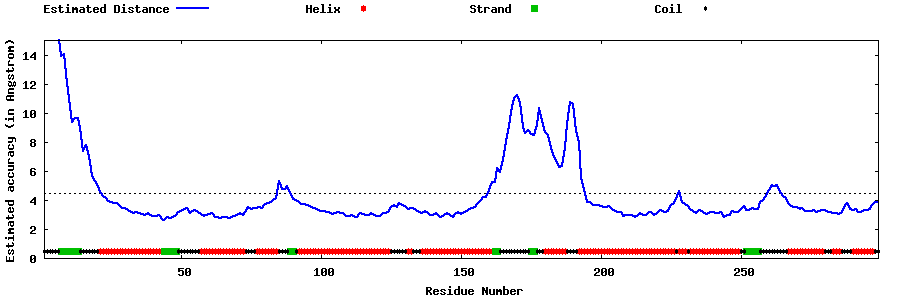 | |||
|
| |||
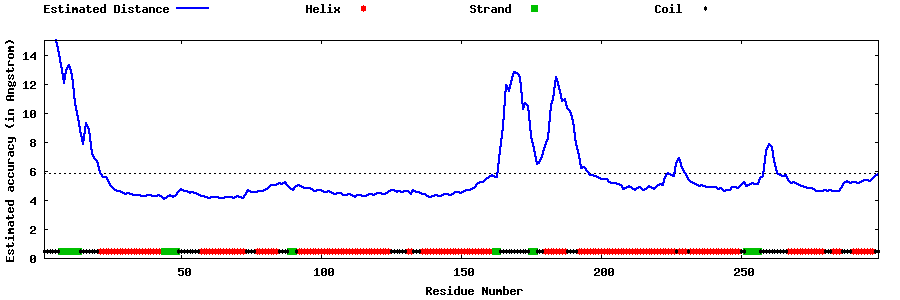 | |||
|
| |||
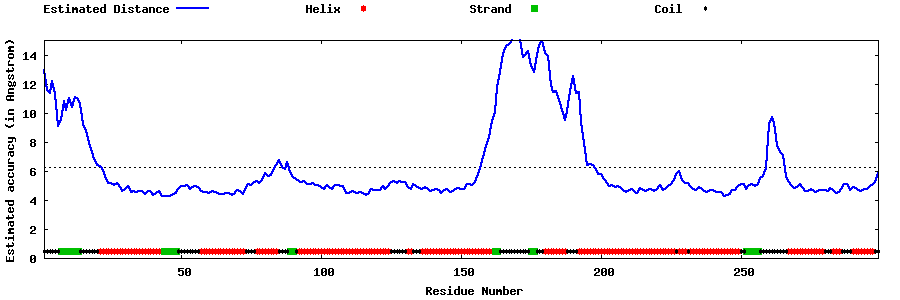 | |||
|
| |||
 | |||
|
| |||