

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MANRNNVTEFILLGLTENPKMQKIIFVVFSVIYINAMIGNVLIVVTITASPSLRSPMYFFLAYLSFIDACYSSVNTPKLITDSLYENKTILFNGCMTQVFGEHFFRGVEVILLTVMAYDHYVAICKPLHYTTVMKQHVCSLLVGVSWVGGFLHATIQILFICQLPFCGPNVIDHFMCDLYTLINLACTNTHTLGLFIAANSGFICLLNCLLLLVSCVVILYSLKTHSLEARHEALSTCVSHITVVILSFIPCIFVYMRPPATLPIDKAVAVFYTMITSMLNPLIYTLRNAQMKNAIRKLCSRKAISSVK | |
| CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCCCSSHHHHHCHHHHCCCHHHCCCCHHHHHHHHHHHHHHCCCCCC | |
| 999873103477258998024799999999999999997887545122079999978999886998764210457589999874399678489999999999999998999999998652887461011610277758999999999999999999999942089889884677643818877783024152201111224599999999999999999999500473021089989988999264663064368868899987501122456430330103643246199999999999711565679 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MANRNNVTEFILLGLTENPKMQKIIFVVFSVIYINAMIGNVLIVVTITASPSLRSPMYFFLAYLSFIDACYSSVNTPKLITDSLYENKTILFNGCMTQVFGEHFFRGVEVILLTVMAYDHYVAICKPLHYTTVMKQHVCSLLVGVSWVGGFLHATIQILFICQLPFCGPNVIDHFMCDLYTLINLACTNTHTLGLFIAANSGFICLLNCLLLLVSCVVILYSLKTHSLEARHEALSTCVSHITVVILSFIPCIFVYMRPPATLPIDKAVAVFYTMITSMLNPLIYTLRNAQMKNAIRKLCSRKAISSVK | |
| 875422000000000042440010001313332332333233000002004304000010023001200110000002000000253210005000000011033113102000200300000002211010000331000001201310230130013102303000112200010022200300002023000300220132023103313311210020013214612220110030021003013210100000033313300000031023103300300002144023002301443134678 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCCHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCSSCCCHHHHHHHHCCCCHHHSSHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHCSSSSSCCCCCCCCCCCSSHHHHHCHHHHCCCHHHCCCCHHHHHHHHHHHHHHCCCCCC MANRNNVTEFILLGLTENPKMQKIIFVVFSVIYINAMIGNVLIVVTITASPSLRSPMYFFLAYLSFIDACYSSVNTPKLITDSLYENKTILFNGCMTQVFGEHFFRGVEVILLTVMAYDHYVAICKPLHYTTVMKQHVCSLLVGVSWVGGFLHATIQILFICQLPFCGPNVIDHFMCDLYTLINLACTNTHTLGLFIAANSGFICLLNCLLLLVSCVVILYSLKTHSLEARHEALSTCVSHITVVILSFIPCIFVYMRPPATLPIDKAVAVFYTMITSMLNPLIYTLRNAQMKNAIRKLCSRKAISSVK | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.20 | 0.89 | 3.43 | Download | ------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYF-----------NFFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.21 | 0.25 | 0.89 | 2.17 | Download | -ENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL--------GWNCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM---------- | |||||||||||||||||||
| 3 | 5tgzA | 0.20 | 0.25 | 0.90 | 2.10 | Download | -GGRGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVC-------------------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAARMDIELAKTLVLILVVLIICWGPLLAIMVGKMNLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 4 | 3uon | 0.17 | 0.21 | 0.88 | 1.55 | Download | ------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF------SN---AAVTFGTAIAAFYLPVIIMTVLYWHISRAKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCANTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.24 | 0.91 | 1.21 | Download | TTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------- | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.20 | 0.89 | 3.51 | Download | -------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARQLVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.22 | 0.85 | 1.74 | Download | -------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQASE----------CVVNT-------D---HILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.22 | 0.87 | 2.81 | Download | --NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGTPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-----------SDEQMLTNALVYVSAAINPILYNLVSANFRQVFLSTL--------- | |||||||||||||||||||
| 9 | 5tgzA | 0.21 | 0.25 | 0.89 | 4.79 | Download | GENFMDIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMN-FAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 10 | 2ydoA | 0.16 | 0.18 | 0.93 | 5.52 | Download | ---------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRLKQMESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQE | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
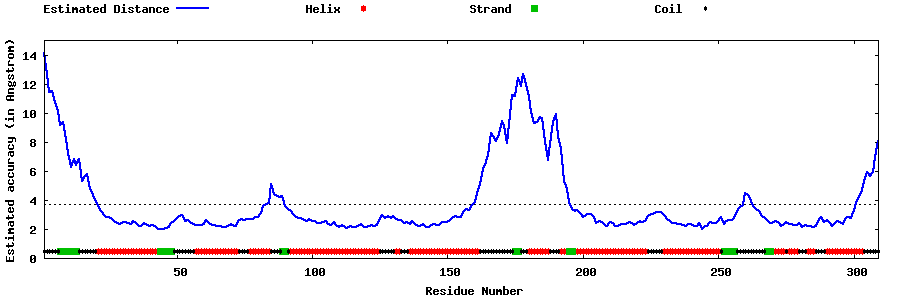 | |||
|
|
|||
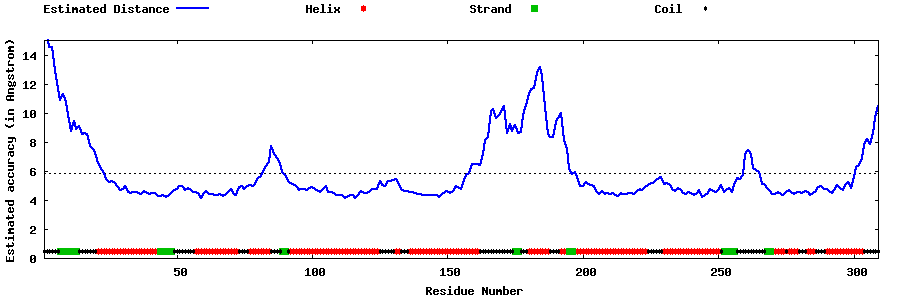 | |||
|
| |||
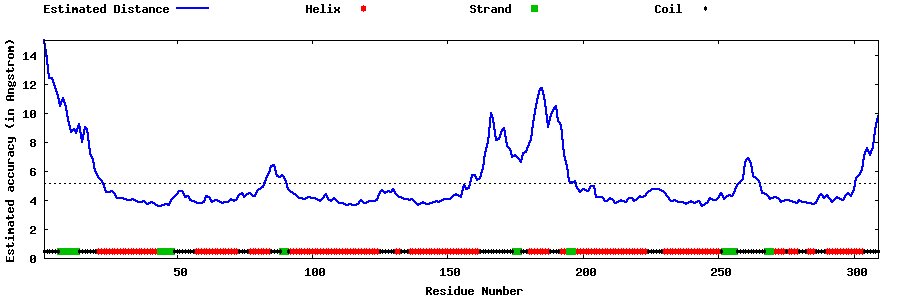 | |||
|
| |||
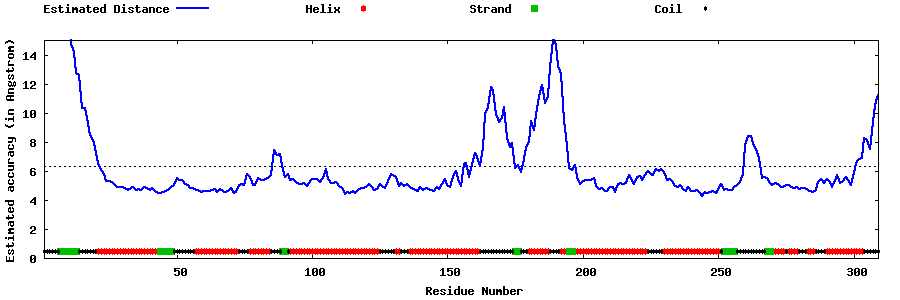 | |||
|
| |||
 | |||
|
| |||