

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MAKNNLTRVTEFILMGFMDHPKLEIPLFLVFLSFYLVTLLGNVGMIMLIQVDVKLYTPMYFFLSHLSLLDACYTSVITPQILATLATGKTVISYGHCAAQFFLFTICAGTECFLLAVMAYDRYAAIRNPLLYTVAMNPRLCWSLVVGAYVCGVSGAILRTTCTFTLSFCKDNQINFFFCDLPPLLKLACSDTANIEIVIIFFGNFVILANASVILISYLLIIKTILKVKSSGGRAKTFSTCASHITAVALFFGALIFMYLQSGSGKSLEEDKVVSVFYTVVIPMLNPLIYSLRNKDVKDAFRKVARRLQVSLSM | |
| CCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHHCCCHHHHHHHHHHHHHHHCCCCC | |
| 99988742314578637898260899999999999999998879999985288877738888767998835721247199989971489788689999999999999999999999997541340162202880267889999999999999999999999984540889892385416818888770568627677788888999999999999999999999802677676654310648889979999743314583689988787897899882210312435644046598999999999864212569 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MAKNNLTRVTEFILMGFMDHPKLEIPLFLVFLSFYLVTLLGNVGMIMLIQVDVKLYTPMYFFLSHLSLLDACYTSVITPQILATLATGKTVISYGHCAAQFFLFTICAGTECFLLAVMAYDRYAAIRNPLLYTVAMNPRLCWSLVVGAYVCGVSGAILRTTCTFTLSFCKDNQINFFFCDLPPLLKLACSDTANIEIVIIFFGNFVILANASVILISYLLIIKTILKVKSSGGRAKTFSTCASHITAVALFFGALIFMYLQSGSGKSLEEDKVVSVFYTVVIPMLNPLIYSLRNKDVKDAFRKVARRLQVSLSM | |
| 85662302001000000043240100001303331331333332012002000300000000010000100100000002000200254220103000000100000000000000000010000002111000300330000001100010010010100000202014422010000012100200013230101000210131133013003303310110001030362231010000002100002320110000003243124441000021032002300300002133024003300432311344 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCHHHCHHHHHHHHHHHHHHHSSSSSCCCCCCCCCCCCSSSSSSCCHHHCCCCHHHHCCCHHHHHHHHHHHHHHHCCCCC MAKNNLTRVTEFILMGFMDHPKLEIPLFLVFLSFYLVTLLGNVGMIMLIQVDVKLYTPMYFFLSHLSLLDACYTSVITPQILATLATGKTVISYGHCAAQFFLFTICAGTECFLLAVMAYDRYAAIRNPLLYTVAMNPRLCWSLVVGAYVCGVSGAILRTTCTFTLSFCKDNQINFFFCDLPPLLKLACSDTANIEIVIIFFGNFVILANASVILISYLLIIKTILKVKSSGGRAKTFSTCASHITAVALFFGALIFMYLQSGSGKSLEEDKVVSVFYTVVIPMLNPLIYSLRNKDVKDAFRKVARRLQVSLSM | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.24 | 0.88 | 3.57 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.20 | 0.87 | 2.04 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL------------------------PLLGWNCEKLQHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM---------- | |||||||||||||||||||
| 3 | 5tjvA | 0.17 | 0.21 | 0.89 | 1.97 | Download | -------ENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHV-FHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSDIFPHID-------------------ETYLMFWIGVTSVLLLFIVYAYMYILWKARAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------- | |||||||||||||||||||
| 4 | 4djh | 0.17 | 0.26 | 0.89 | 1.53 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------- | |||||||||||||||||||
| 5 | 4yay | 0.15 | 0.26 | 0.90 | 1.26 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIH-RNVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--------- | |||||||||||||||||||
| 6 | 3emlA | 0.17 | 0.24 | 0.88 | 3.77 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.21 | 0.84 | 1.71 | Download | --------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------ | |||||||||||||||||||
| 8 | 2z73A | 0.16 | 0.19 | 0.95 | 2.92 | Download | W--YNPSIVVHPHWREFDQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTLE------GVLCDY-------ISRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQF | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.24 | 0.88 | 5.25 | Download | ----------------------ISSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-- | |||||||||||||||||||
| 10 | 2ydoA | 0.15 | 0.24 | 0.92 | 6.04 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQE | |||||||||||||||||||
| ||||||||||||||||||||||||||
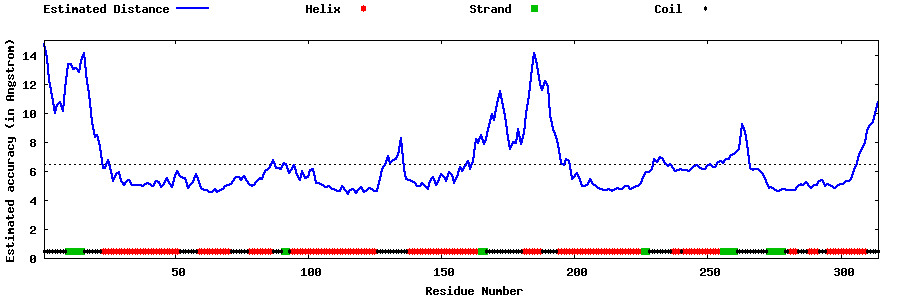
| Generated 3D models | Estimated local accuracy of models | ||
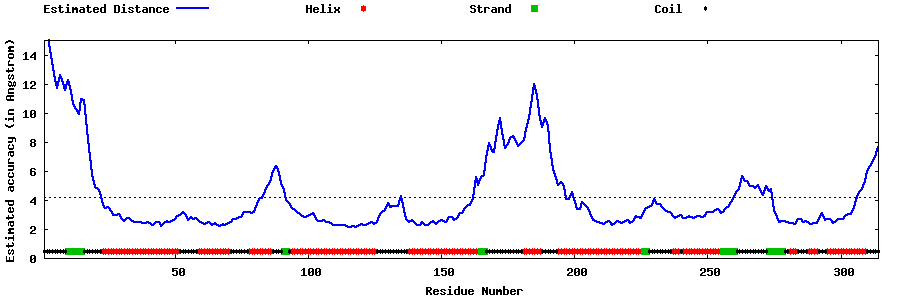 | |||
|
|
|||
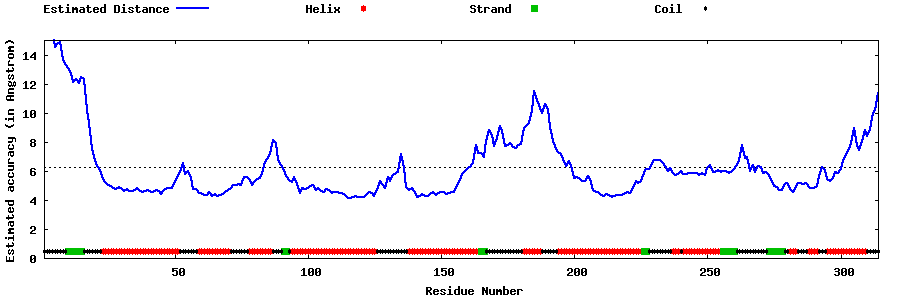 | |||
|
| |||
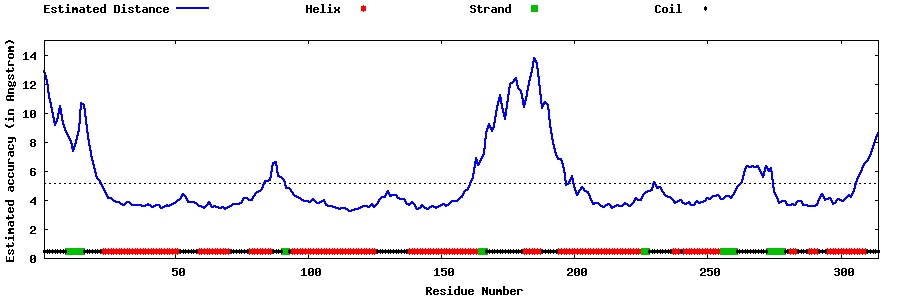 | |||
|
| |||
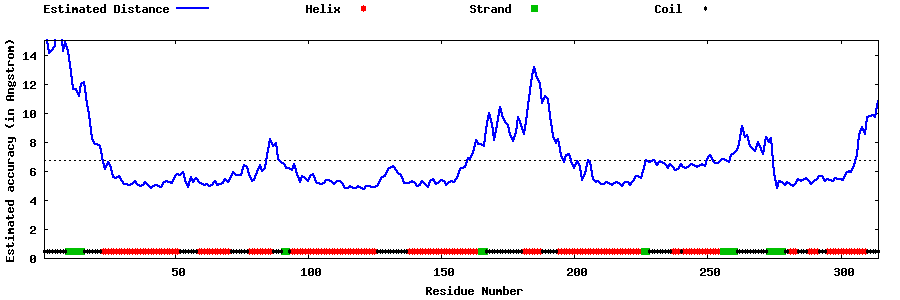 | |||
|
| |||
 | |||
|
| |||