

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| METKNYSSSTSGFILLGLSSNPKLQKPLFAIFLIMYLLTAVGNVLIILAIYSDPRLHTPMYFFLSNLSFMDICFTTVIVPKMLVNFLSETKIISYVGCLIQMYFFMAFGNTDSYLLASMAIDRLVAICNPLHYDVVMKPWHCLLMLLGSCSISHLHSLFRVLLMSRLSFCASHIIKHFFCDTQPVLKLSCSDTSSSQMVVMTETLAVIVTPFLCTIFSYLQIIVTVLRIPSAAGKWKAFSTCGSHLTVVVLFYGSVIYVYFRPLSMYSVMKGRVATVMYTVVTPMLNPFIYSLRNKDMKRGLKKLRHRIYS | |
| CCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCCHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCC | |
| 98978871026770417998831678999999999999999899999999738865563999988799874451316879988998438966758999999999999999999999999886176516543177412771899999999999999999999998751489999227926785898765265436889999999999999979999999999999850045778867221570878999999867031467278987889888189984016562444564236628899999999853079 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| METKNYSSSTSGFILLGLSSNPKLQKPLFAIFLIMYLLTAVGNVLIILAIYSDPRLHTPMYFFLSNLSFMDICFTTVIVPKMLVNFLSETKIISYVGCLIQMYFFMAFGNTDSYLLASMAIDRLVAICNPLHYDVVMKPWHCLLMLLGSCSISHLHSLFRVLLMSRLSFCASHIIKHFFCDTQPVLKLSCSDTSSSQMVVMTETLAVIVTPFLCTIFSYLQIIVTVLRIPSAAGKWKAFSTCGSHLTVVVLFYGSVIYVYFRPLSMYSVMKGRVATVMYTVVTPMLNPFIYSLRNKDMKRGLKKLRHRIYS | |
| 85562450200100001015427212000110030122023312000000000120000000000010110111200310300101115344031300000110010002100000000011000000100200010013000000020020002001000000100010372401000012320030000102101010223133323323310330101000000102146232100000000000000111001000000112324432200000001312330020000103102300230033248 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCHHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHCHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCSSSCCHHHHHHHHHHHCCHHSSSSCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCC METKNYSSSTSGFILLGLSSNPKLQKPLFAIFLIMYLLTAVGNVLIILAIYSDPRLHTPMYFFLSNLSFMDICFTTVIVPKMLVNFLSETKIISYVGCLIQMYFFMAFGNTDSYLLASMAIDRLVAICNPLHYDVVMKPWHCLLMLLGSCSISHLHSLFRVLLMSRLSFCASHIIKHFFCDTQPVLKLSCSDTSSSQMVVMTETLAVIVTPFLCTIFSYLQIIVTVLRIPSAAGKWKAFSTCGSHLTVVVLFYGSVIYVYFRPLSMYSVMKGRVATVMYTVVTPMLNPFIYSLRNKDMKRGLKKLRHRIYS | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.21 | 0.89 | 3.62 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.18 | 0.21 | 0.91 | 2.08 | Download | -GRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-----------------GWNCEKLQSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSMF----- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.21 | 0.87 | 2.07 | Download | -------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-----------------PPFFWRQ----ASECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKYLLMERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.23 | 0.90 | 1.54 | Download | --------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP--- | |||||||||||||||||||
| 5 | 4yay | 0.19 | 0.23 | 0.91 | 1.23 | Download | KTTRN--AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHR-NVFF------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIQIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.21 | 0.88 | 3.83 | Download | ----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.22 | 0.85 | 1.71 | Download | ---------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPF-FWRQAS----------ECVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF--- | |||||||||||||||||||
| 8 | 2rh1A | 0.16 | 0.21 | 0.87 | 2.75 | Download | --------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM----HWYRATHQEAINCYAEE----TCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDE-------------GLIGHLLTKSPSLNAAKSELDKAIGRNTNGVITKDEAEKLFNQDVDAAVRGILRNAKL | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.21 | 0.89 | 4.97 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.19 | 0.92 | 5.62 | Download | ------------------S------SVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFAINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| ||||||||||||||||||||||||||
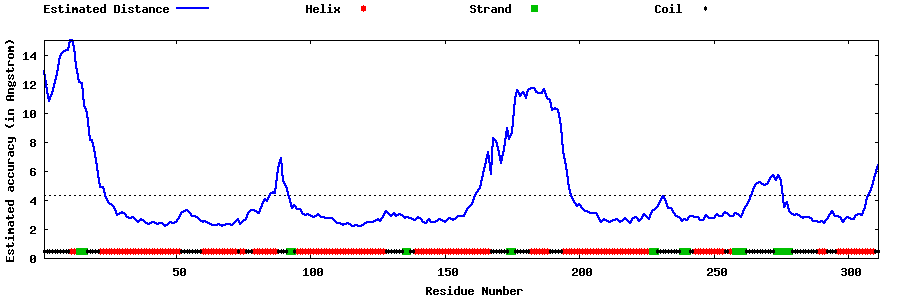
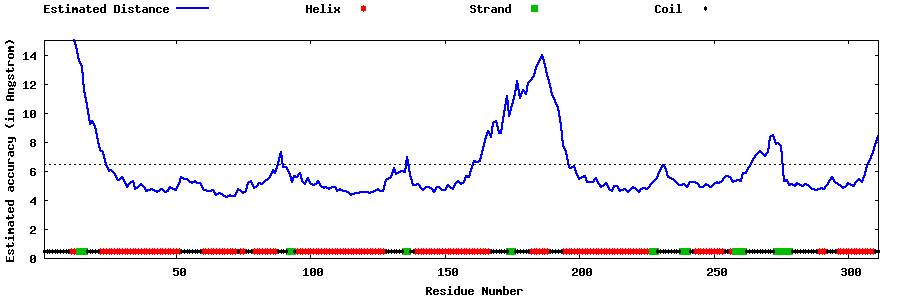
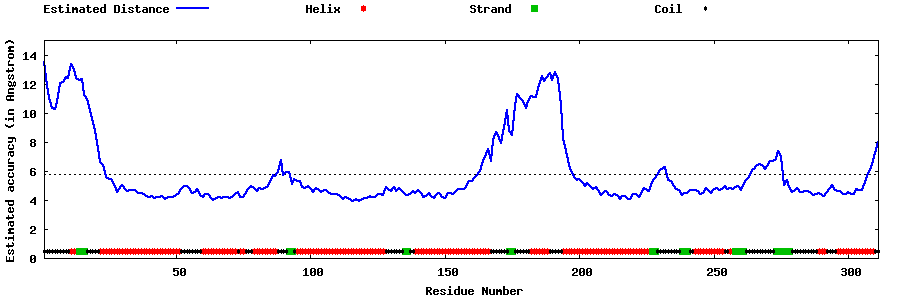
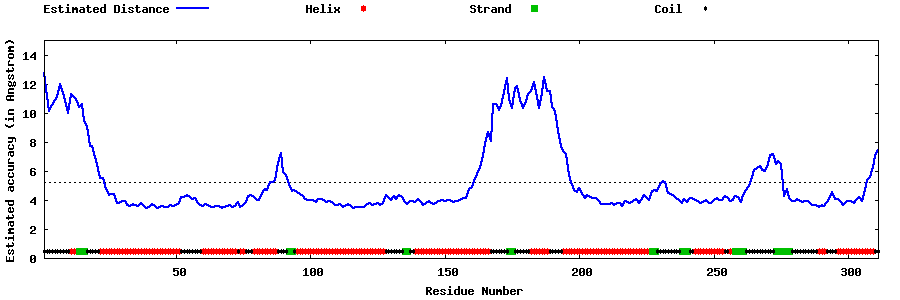
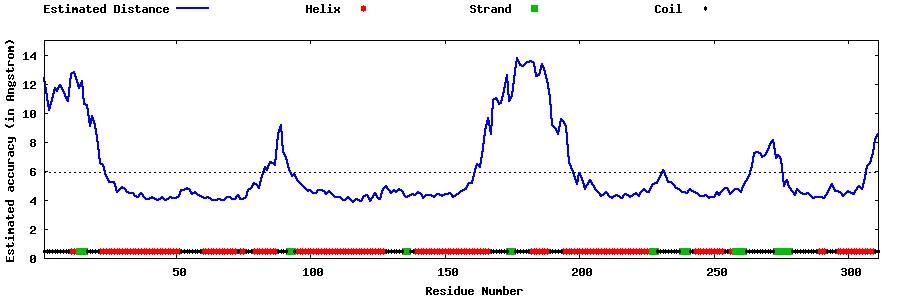
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||