

| 20 40 60 80 100 120 140 160 180 200 220 240 260 | |
| | | | | | | | | | | | | | | |
| MRRKNLTEVTEFVFLGFSRFHKHHITLFVVFLILYTLTVAGNAIIMTIICIDRHLHTPMYFFLSMLASSKTVYTLFIIPQMLSSFVTQTQPISLAGCTTQTFFFVTLAINNCFLLTVMGYDHYMAICNPLRYRVITSKKVCVQLVCGAFSIGLAMAAVQVTSIFTLPFCHTVVGHFFCDILPVMKLSCINTTINEIINFVVRLFVILVPMGLVFISYVLIISTVLKIASAEGWKKTFATCAFHLTVVIVHYGCASIAYLMPKSENSIEQDLLLSVT | |
| CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCSSCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSC | |
| 999887345677776689984349999999999999999989999899972887657488998779999788883377889898656996786899999999999999999999999985511502665448850478799999999999999999999998658227899267654580888888605861999999999999999979999999999999981287736564054456878997999973242268378999988778677519 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 | |
| | | | | | | | | | | | | | | |
| MRRKNLTEVTEFVFLGFSRFHKHHITLFVVFLILYTLTVAGNAIIMTIICIDRHLHTPMYFFLSMLASSKTVYTLFIIPQMLSSFVTQTQPISLAGCTTQTFFFVTLAINNCFLLTVMGYDHYMAICNPLRYRVITSKKVCVQLVCGAFSIGLAMAAVQVTSIFTLPFCHTVVGHFFCDILPVMKLSCINTTINEIINFVVRLFVILVPMGLVFISYVLIISTVLKIASAEGWKKTFATCAFHLTVVIVHYGCASIAYLMPKSENSIEQDLLLSVT | |
| 875623020000000000432501010013033313312333320020010133000000210210022032112033030002011663201030022012001000210010001001100000021120213003300000021012101310310011003020143301000002210030003203110221122123313313310330332000000103246323200000000200000112000000010326335434103207 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCSSCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSSCCCCCCCCCSSCCHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHSSSSCCCCCCCCCCCSSSSSC MRRKNLTEVTEFVFLGFSRFHKHHITLFVVFLILYTLTVAGNAIIMTIICIDRHLHTPMYFFLSMLASSKTVYTLFIIPQMLSSFVTQTQPISLAGCTTQTFFFVTLAINNCFLLTVMGYDHYMAICNPLRYRVITSKKVCVQLVCGAFSIGLAMAAVQVTSIFTLPFCHTVVGHFFCDILPVMKLSCINTTINEIINFVVRLFVILVPMGLVFISYVLIISTVLKIASAEGWKKTFATCAFHLTVVIVHYGCASIAYLMPKSENSIEQDLLLSVT | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.19 | 0.87 | 2.94 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIS | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.21 | 0.80 | 1.91 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL-----------------------PLLGWNCEKLQSVDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICW------------------------- | |||||||||||||||||||
| 3 | 4iaqA | 0.13 | 0.21 | 0.89 | 2.07 | Download | ------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISILYTVYSTVGAF-----------YFPTLLLIALYGRIYVEARSRKATKTLGIILGAFIVCWLPFFIISLVMPHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 4 | 3d4s | 0.15 | 0.16 | 0.89 | 1.52 | Download | ----------------------WVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWYRA---THQEAINCYAEETC---CDFF-TNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNILKEHKALKTLGIIMGTFTLCWLPFFIVNIVHVIQDNLIRKEVYILL | |||||||||||||||||||
| 5 | 4yay | 0.15 | 0.22 | 0.93 | 1.27 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVF-F-----IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIRDCRIDTAMPIT | |||||||||||||||||||
| 6 | 3emlA | 0.17 | 0.19 | 0.87 | 3.18 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIV | |||||||||||||||||||
| 7 | 4iaq | 0.16 | 0.24 | 0.67 | 1.69 | Download | --------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQAS---------ECVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARS-------------------------------------------------- | |||||||||||||||||||
| 8 | 2rh1A | 0.17 | 0.16 | 0.72 | 2.23 | Download | -------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQM------------HWYCD-----------FFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKRQLNIFEMLRIDEG------------------------------------L | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.19 | 0.88 | 4.14 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIV | |||||||||||||||||||
| 10 | 2ydoA | 0.14 | 0.18 | 0.91 | 5.07 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLTFFCPDCSHAPLWLMYLAIV | |||||||||||||||||||
| ||||||||||||||||||||||||||
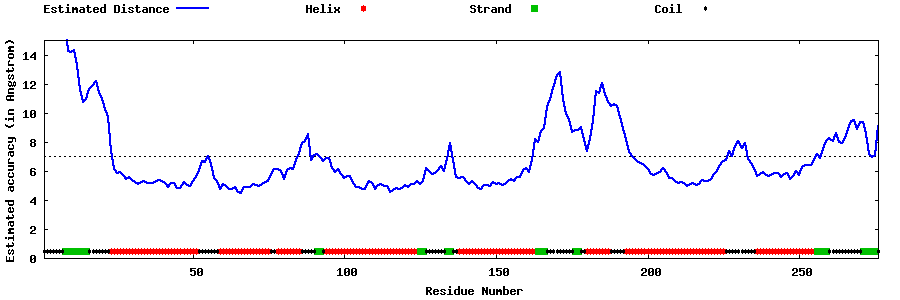
| Generated 3D models | Estimated local accuracy of models | ||
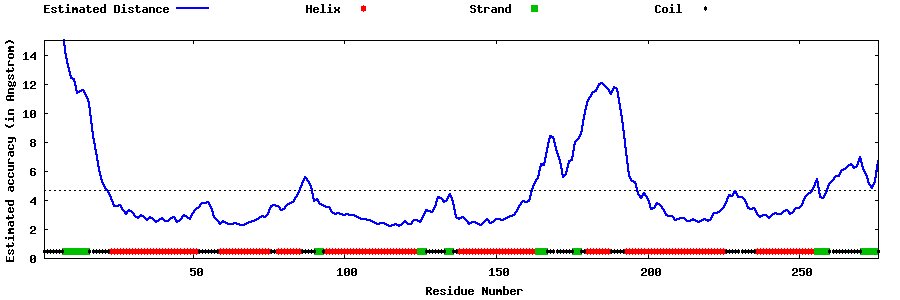 | |||
|
|
|||
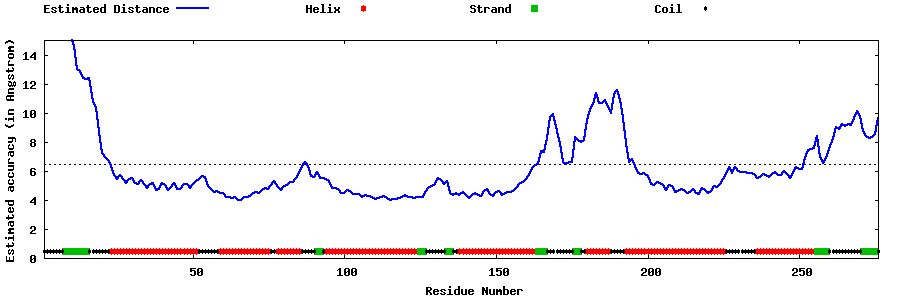 | |||
|
| |||
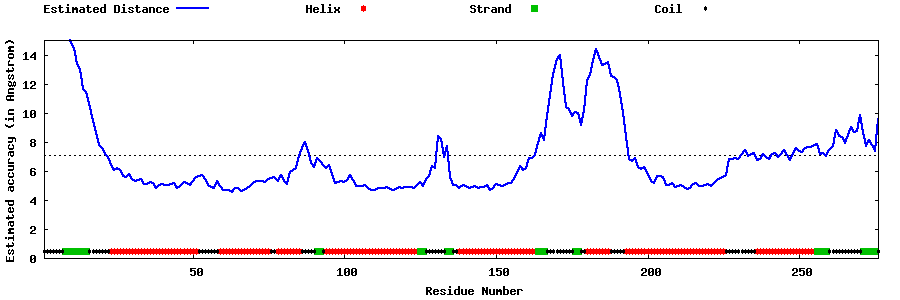 | |||
|
| |||
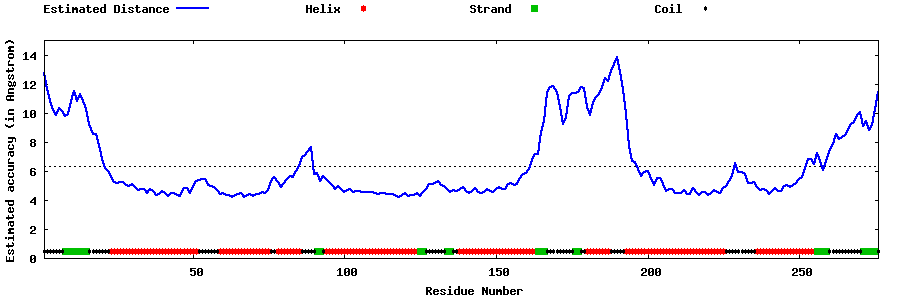 | |||
|
| |||
 | |||
|
| |||