

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MANITRMANHTGRLDFILMGLFRQSKHPALLSVVIFVVFLKALSGNAVLILLIHCDAHLHSPMYFFISQLSLMDMAYISVTVPKMLLDQVMGVNKVSAPECGMQMFLYLTLAGSEFFLLATMAYDRYVAICHPLRYPVLMNHRVCLFLASGCWFLGSVDGFMLTPITMSFPFCRSWEIHHFFCEVPAVTILSCSDTSLYETLMYLCCVLMLLIPVTIISSSYLLILLTVHRMNSAEGRKKAFATCSSHLTVVILFYGAAVYTYMLPSSYHTPEKDMMVSVFYTILTPVLNPLIYSLRNKDVMGALKKMLTVRFVL | |
| CCCHHHCCCCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCC | |
| 901231666753204578779998224889999999999999998999989997388755639999888999988877115899999850599588689999999999999999999999998651462055556884248869999999999999999999999990517899795787514838888886147519999999999999999799999999999999833876121541765178789989999865774251899999987785897416327754024533146499999999997422089 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MANITRMANHTGRLDFILMGLFRQSKHPALLSVVIFVVFLKALSGNAVLILLIHCDAHLHSPMYFFISQLSLMDMAYISVTVPKMLLDQVMGVNKVSAPECGMQMFLYLTLAGSEFFLLATMAYDRYVAICHPLRYPVLMNHRVCLFLASGCWFLGSVDGFMLTPITMSFPFCRSWEIHHFFCEVPAVTILSCSDTSLYETLMYLCCVLMLLIPVTIISSSYLLILLTVHRMNSAEGRKKAFATCSSHLTVVILFYGAAVYTYMLPSSYHTPEKDMMVSVFYTILTPVLNPLIYSLRNKDVMGALKKMLTVRFVL | |
| 564135341303211000000033361210001203332331333332002001001300000011020000101111002102000000154320001002201100110111002000300200000002102000200330000001100310031031000100001003423010000022100300020033102101331333133123203301330010001031461231010000001000001320000000103243346311000011023203300300001042023002200333126 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHCCCCCCCCSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSSHHHHHHCCCCHHHCCCCHHHHHHHHHHHHHCCCC MANITRMANHTGRLDFILMGLFRQSKHPALLSVVIFVVFLKALSGNAVLILLIHCDAHLHSPMYFFISQLSLMDMAYISVTVPKMLLDQVMGVNKVSAPECGMQMFLYLTLAGSEFFLLATMAYDRYVAICHPLRYPVLMNHRVCLFLASGCWFLGSVDGFMLTPITMSFPFCRSWEIHHFFCEVPAVTILSCSDTSLYETLMYLCCVLMLLIPVTIISSSYLLILLTVHRMNSAEGRKKAFATCSSHLTVVILFYGAAVYTYMLPSSYHTPEKDMMVSVFYTILTPVLNPLIYSLRNKDVMGALKKMLTVRFVL | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.22 | 0.88 | 3.50 | Download | ------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 2 | 5tgzA | 0.19 | 0.22 | 0.89 | 2.27 | Download | -----RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKLQSC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAMLCLLNSTVNPIIYALRSKDLRHAFRSM------- | |||||||||||||||||||
| 3 | 4iaqA | 0.17 | 0.19 | 0.83 | 2.24 | Download | ----------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL---------PPCVVNTDH-----------------ILYTVYSTVGAFY---FPTLLLIALYGRIYVEARSQKYLLMAARERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 4 | 4djh | 0.16 | 0.24 | 0.89 | 1.53 | Download | -----------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVR---EDVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---- | |||||||||||||||||||
| 5 | 4yay | 0.16 | 0.23 | 0.91 | 1.24 | Download | AEQLKTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLQLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL------ | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.22 | 0.88 | 3.76 | Download | -------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.19 | 0.83 | 1.71 | Download | --------------------------WKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK--- | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.17 | 0.90 | 2.81 | Download | --------NSD-------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGPI----------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDEQWTNALVYVSAAINPILYNLVSANFRQVFLSTL------ | |||||||||||||||||||
| 9 | 3emlA | 0.19 | 0.22 | 0.88 | 4.61 | Download | ------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAK-SLAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.19 | 0.91 | 5.39 | Download | ---------------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLR | |||||||||||||||||||
| ||||||||||||||||||||||||||
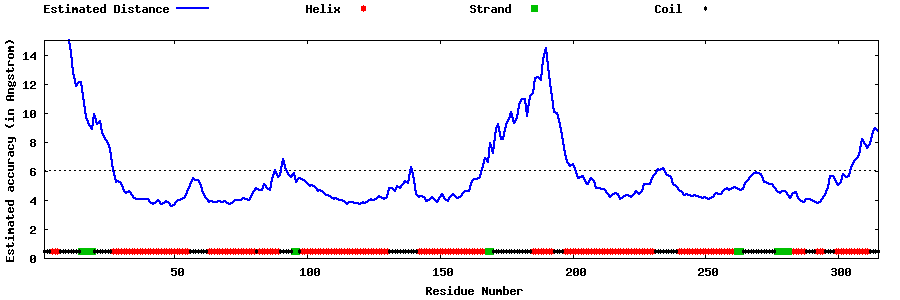
| Generated 3D models | Estimated local accuracy of models | ||
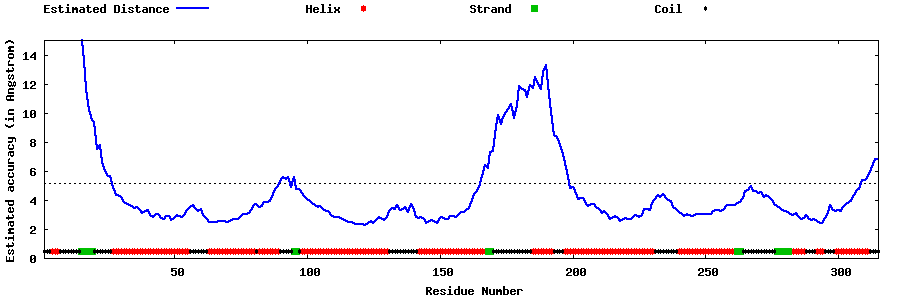 | |||
|
|
|||
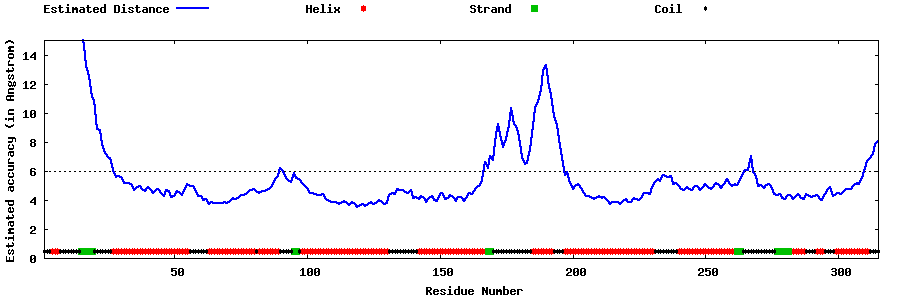 | |||
|
| |||
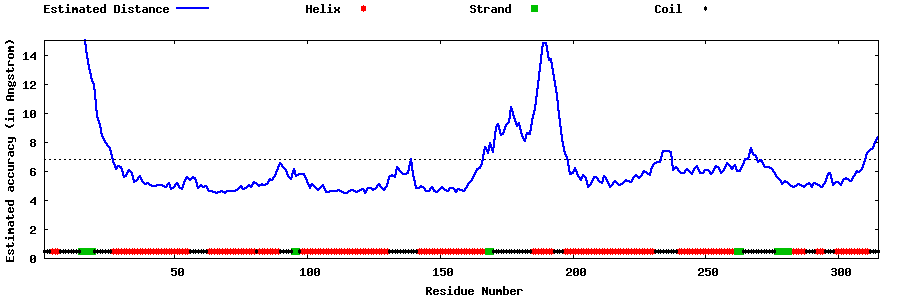 | |||
|
| |||
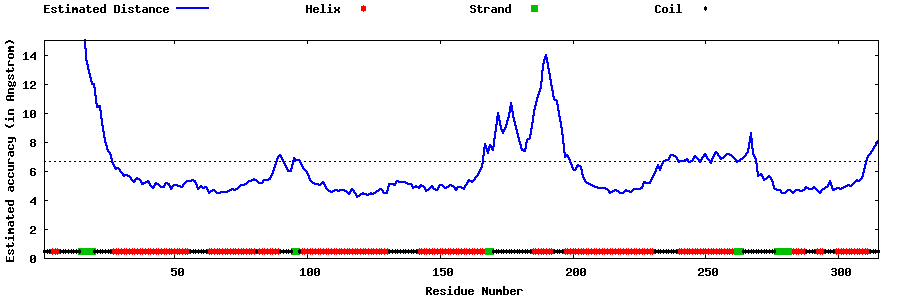 | |||
|
| |||
 | |||
|
| |||