

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MPLFNSLCWFPTIHVTPPSFILNGIPGLERVHVWISLPLCTMYIIFLVGNLGLVYLIYYEESLHHPMYFFFGHALSLIDLLTCTTTLPNALCIFWFSLKEINFNACLAQMFFVHGFTGVESGVLMLMALDRYVAICYPLRYATTLTNPIIAKAELATFLRGVLLMIPFPFLVKRLPFCQSNIISHTYCDHMSVVKLSCASIKVNVIYGLMVALLIGVFDICCISLSYTLILKAAISLSSSDARQKAFSTCTAHISAIIITYVPAFFTFFAHRFGGHTIPPSLHIIVANLYLLLPPTLNPIVYGVKTKQIRKSVIKFFQGDKGAG | |
| CCCCCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHCCCCCC | |
| 997564423689987552114779999866799999999999999999999999998647875426699999899999999998569999999826998788878999999999999999999999980316862665457602868999999999999999999889999825899998968873320066788843794154599999999999999999999999999998078898898999772899999999999999999500045888899336689999985277553570110556999999999971266899 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | |
| | | | | | | | | | | | | | | | | | |
| MPLFNSLCWFPTIHVTPPSFILNGIPGLERVHVWISLPLCTMYIIFLVGNLGLVYLIYYEESLHHPMYFFFGHALSLIDLLTCTTTLPNALCIFWFSLKEINFNACLAQMFFVHGFTGVESGVLMLMALDRYVAICYPLRYATTLTNPIIAKAELATFLRGVLLMIPFPFLVKRLPFCQSNIISHTYCDHMSVVKLSCASIKVNVIYGLMVALLIGVFDICCISLSYTLILKAAISLSSSDARQKAFSTCTAHISAIIITYVPAFFTFFAHRFGGHTIPPSLHIIVANLYLLLPPTLNPIVYGVKTKQIRKSVIKFFQGDKGAG | |
| 644322112213343311102000001213202000110120122023212100000201510010001000300010000002000020000000204403040000001200310130100001002000000021010000002410010000003212011202100021022044200000001021001000230300011002003313332231032011100200020024601320120010010001003231331010000142400110002102310030033003000020340041003002444778 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHHCCCCCC MPLFNSLCWFPTIHVTPPSFILNGIPGLERVHVWISLPLCTMYIIFLVGNLGLVYLIYYEESLHHPMYFFFGHALSLIDLLTCTTTLPNALCIFWFSLKEINFNACLAQMFFVHGFTGVESGVLMLMALDRYVAICYPLRYATTLTNPIIAKAELATFLRGVLLMIPFPFLVKRLPFCQSNIISHTYCDHMSVVKLSCASIKVNVIYGLMVALLIGVFDICCISLSYTLILKAAISLSSSDARQKAFSTCTAHISAIIITYVPAFFTFFAHRFGGHTIPPSLHIIVANLYLLLPPTLNPIVYGVKTKQIRKSVIKFFQGDKGAG | |||||||||||||||||||||||||
| 1 | 3emlA | 0.15 | 0.20 | 0.86 | 2.28 | Download | ---------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQN-VTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.24 | 0.83 | 2.09 | Download | -----------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGS-LAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 3 | 5tgzA | 0.21 | 0.24 | 0.88 | 1.81 | Download | GGRGENFMDIECFMVLNP-----------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSY-HFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSD-IFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.23 | 0.87 | 1.53 | Download | -------------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLM-SLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKG-IET----N-PNNITCVLTK-------E--RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADSNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-- | |||||||||||||||||||
| 5 | 3uonA | 0.17 | 0.19 | 0.84 | 1.12 | Download | ----------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLF-SLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQF--IVGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTV-WTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------ | |||||||||||||||||||
| 6 | 3emlA | 0.16 | 0.20 | 0.85 | 2.54 | Download | -----------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVV-SLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 3d4s | 0.16 | 0.17 | 0.60 | 1.70 | Download | --------------------------------VGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITS-LACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWY----RATHQEAINCYAE----ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKR---------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.18 | 0.18 | 0.89 | 2.96 | Download | ----------------------NDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGS-LALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIV------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------- | |||||||||||||||||||
| 9 | 5tgzA | 0.20 | 0.24 | 0.89 | 3.00 | Download | -----GGRGENFMDI--ECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 10 | 4gpoA | 0.15 | 0.19 | 0.86 | 4.44 | Download | ---------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNL-FITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP-DWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL-------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
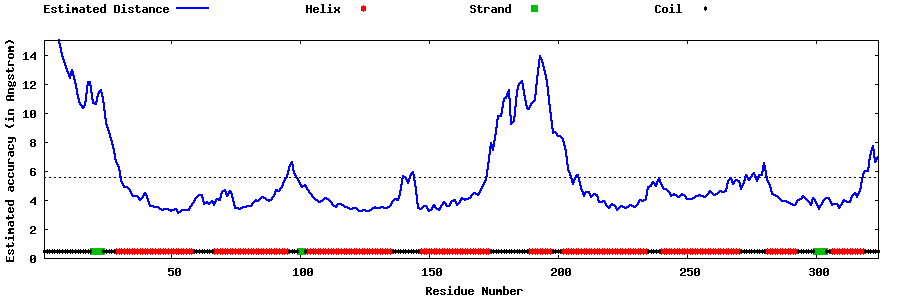
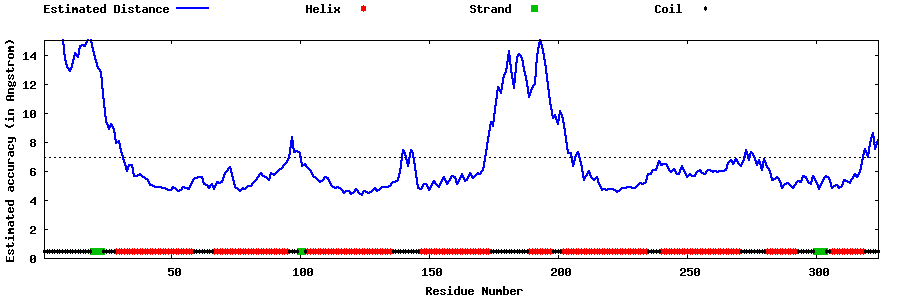
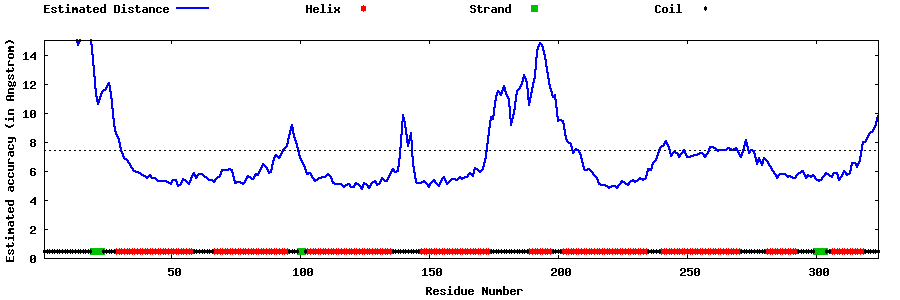
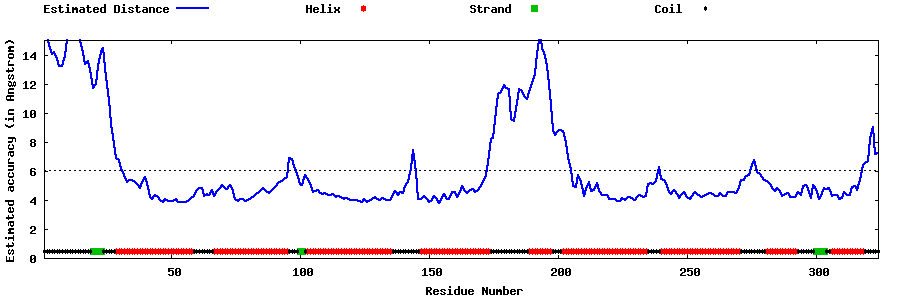
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||