

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MTETSLSSQCFPMSVLNNTIAEPLIFLLMGIPGLKATQYWISIPFCLLYVVAVSGNSMILFVVLCERSLHKPMYYFLSMLSATDLSLSLCTLSTTLGVFWFEAREINLNACIAQMFFLHGFTFMESGVLLAMAFDRFVAICYPLRYTTILTNARIAKIGMSMLIRNVAVMLPVMLFVKRLSFCSSMVLSHSYCYHVDLIQLSCTDNRINSILGLFALLSTTGFDCPCILLSYILIIRSVLSIASSEERRKAFNTCTSHISAVSIFYLPLISLSLVHRYGHSAPPFVHIIMANVFLLIPPVLNPIIYSVKIKQIQKAIIKVLIQKHSKSNHQLFLIRDKAIYE | |
| CCCCCCCCCCCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCCCCCSSSSCCCHHCCC | |
| 986555767676888789988563002689989855779999999999999999999999988578875422899999999999879987799999999659986888789999999999999999999999823055216665563038789999999999999999998899999528999999547723203767888067836768999999999999989999999999999983688876888986405999999999999899997464325888826999999999838766478333154799999999996167788765244201010249 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | |
| | | | | | | | | | | | | | | | | | | |
| MTETSLSSQCFPMSVLNNTIAEPLIFLLMGIPGLKATQYWISIPFCLLYVVAVSGNSMILFVVLCERSLHKPMYYFLSMLSATDLSLSLCTLSTTLGVFWFEAREINLNACIAQMFFLHGFTFMESGVLLAMAFDRFVAICYPLRYTTILTNARIAKIGMSMLIRNVAVMLPVMLFVKRLSFCSSMVLSHSYCYHVDLIQLSCTDNRINSILGLFALLSTTGFDCPCILLSYILIIRSVLSIASSEERRKAFNTCTSHISAVSIFYLPLISLSLVHRYGHSAPPFVHIIMANVFLLIPPVLNPIIYSVKIKQIQKAIIKVLIQKHSKSNHQLFLIRDKAIYE | |
| 645352444323023213142313102000001213202000110120122023212100000201410010001000000200100030010200000002045030400000012003102301100010020000000210100000023100100000022120112021010210210452000000010100010002303000110010033133332310320111002000100146013201100100000000032113210100100331011000000220003003300300002033004100300133645442202024564348 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCCCCCCCCCCCCSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCCCCCSSSSCCCHHCCC MTETSLSSQCFPMSVLNNTIAEPLIFLLMGIPGLKATQYWISIPFCLLYVVAVSGNSMILFVVLCERSLHKPMYYFLSMLSATDLSLSLCTLSTTLGVFWFEAREINLNACIAQMFFLHGFTFMESGVLLAMAFDRFVAICYPLRYTTILTNARIAKIGMSMLIRNVAVMLPVMLFVKRLSFCSSMVLSHSYCYHVDLIQLSCTDNRINSILGLFALLSTTGFDCPCILLSYILIIRSVLSIASSEERRKAFNTCTSHISAVSIFYLPLISLSLVHRYGHSAPPFVHIIMANVFLLIPPVLNPIIYSVKIKQIQKAIIKVLIQKHSKSNHQLFLIRDKAIYE | |||||||||||||||||||||||||
| 1 | 3emlA | 0.18 | 0.19 | 0.81 | 2.33 | Download | ---------------------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------------- | |||||||||||||||||||
| 2 | 5tgzA | 0.20 | 0.22 | 0.79 | 2.05 | Download | -----------------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------- | |||||||||||||||||||
| 3 | 4zwjA | 0.16 | 0.23 | 0.96 | 1.87 | Download | AYMCGTEGPNFYVPFSNATGVVRSPFEYPQYYLAPWQFSMLAAYMFLLIVLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVLGGFTSTLYTSLHGYFVFGPTGCNLQGFFATLGGEIALWSLVVLAIERYVVVCKPM-SNFRFGENHAIMGVAFTWVMALACAAP-------PLAGWSRYIPEGLQ-------CSCGIDYYNESFVIYMFVVHFTIPMIIIFFCYGQLVFTVKEAATQKAEKEVTRMVIIYVIAFLICWVPYASVAFYIFTHQGSFGPIFMTIPAFFAKSAAIYNPVIYIMMNKQFRNCMLTTICKKVSRDKSVTIYLGKRDYVD | |||||||||||||||||||
| 4 | 4djh | 0.15 | 0.21 | 0.82 | 1.49 | Download | ---------------------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVTT-TMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVRE---DVDVIECSLQFPDD---DYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSA--ALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP------------------- | |||||||||||||||||||
| 5 | 3uonA | 0.17 | 0.17 | 0.80 | 1.12 | Download | ----------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQFF---------SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM-------------------- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.19 | 0.81 | 2.53 | Download | -----------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC---PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ-------------- | |||||||||||||||||||
| 7 | 3d4s | 0.16 | 0.23 | 0.57 | 1.70 | Download | -------------------------------------VVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIWTLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTSFLPIQMHWY----RATHQEAINCYAE----ETCCDFFTNQAYAIASSIVSFYVPLVIMVFVYSRVFQEAKR----------------------------------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4buoA | 0.16 | 0.18 | 0.85 | 3.03 | Download | ----------------NSDL---------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGGLVCTPIV------DTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL--------------------- | |||||||||||||||||||
| 9 | 5tgzA | 0.19 | 0.22 | 0.83 | 3.05 | Download | -----------GGRGENFMDI--ECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--------------------- | |||||||||||||||||||
| 10 | 4gpoA | 0.17 | 0.17 | 0.81 | 4.22 | Download | ---------------------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVPDWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL---------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
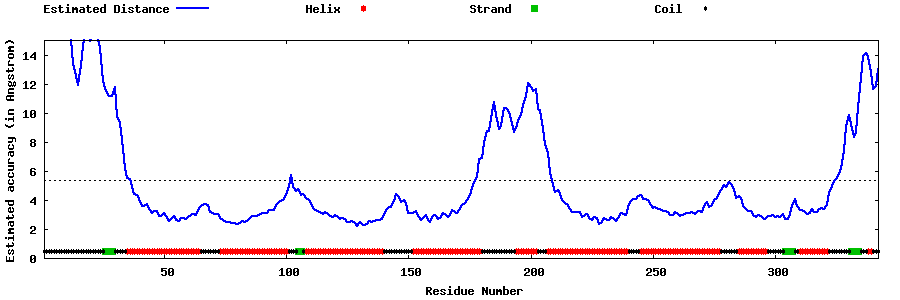 | |||
|
|
|||
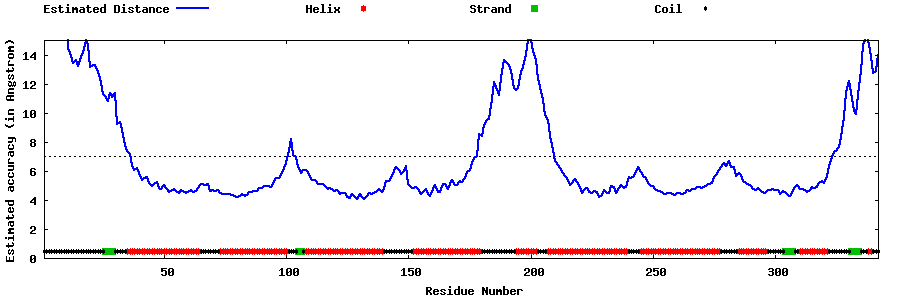 | |||
|
| |||
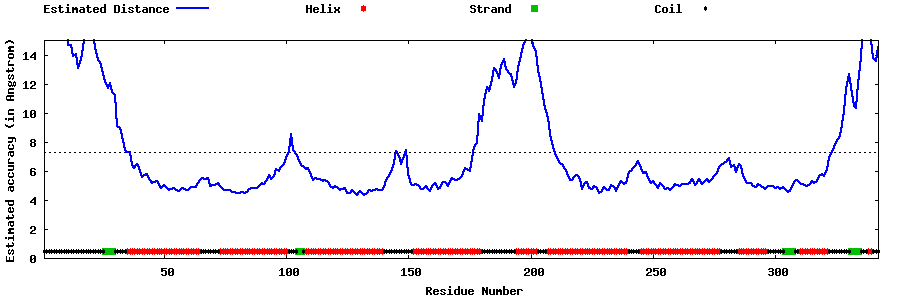 | |||
|
| |||
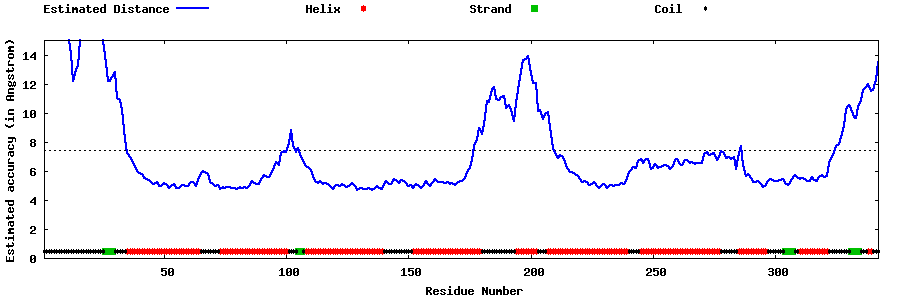 | |||
|
| |||
 | |||
|
| |||