

| 20 40 60 80 100 120 140 | |
| | | | | | | | | |
| MVSANQTASVTEFILLGLSAHPKLEKTFFVLILLMYLVILLGNGVLILMTVSNSHLHMPMYFFLGNLSFLDICYTTSSVPLILDSFLTPRKTISFSACAVQMFLSFAMGATECVLLSMMAFDRYVAICNPLRYPVVMSKAAYMPIRLPAPG | |
| CCCCCCCSSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHCC | |
| 9987885344233565589985079999999999999999988999899970887667589995679999888870578899998647996572898899999999999999999999987518871655447600588899988813069 |
| 20 40 60 80 100 120 140 | |
| | | | | | | | | |
| MVSANQTASVTEFILLGLSAHPKLEKTFFVLILLMYLVILLGNGVLILMTVSNSHLHMPMYFFLGNLSFLDICYTTSSVPLILDSFLTPRKTISFSACAVQMFLSFAMGATECVLLSMMAFDRYVAICNPLRYPVVMSKAAYMPIRLPAPG | |
| 8566444230220000000633512221123033323323333330120022244111001220332012011322233131032022653302030033023313311331130013003210000032231221023300110023028 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHCC MVSANQTASVTEFILLGLSAHPKLEKTFFVLILLMYLVILLGNGVLILMTVSNSHLHMPMYFFLGNLSFLDICYTTSSVPLILDSFLTPRKTISFSACAVQMFLSFAMGATECVLLSMMAFDRYVAICNPLRYPVVMSKAAYMPIRLPAPG | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.24 | 0.85 | 2.12 | Download | ---------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWV | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.22 | 0.85 | 1.76 | Download | --------------------NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMW- | |||||||||||||||||||
| 3 | 4iaqA | 0.18 | 0.17 | 0.91 | 1.95 | Download | -------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWV | |||||||||||||||||||
| 4 | 4iaq | 0.19 | 0.17 | 0.91 | 1.52 | Download | -------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALLPF | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.26 | 0.99 | 1.23 | Download | LKTTRN-AYIQKYLILNSSDCPYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWL | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.24 | 0.84 | 2.26 | Download | ----------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWV | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.17 | 0.85 | 1.71 | Download | ---------------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVW- | |||||||||||||||||||
| 8 | 2rh1A | 0.21 | 0.18 | 0.87 | 1.88 | Download | --------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILMKMWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWI | |||||||||||||||||||
| 9 | 5tgzA | 0.20 | 0.22 | 0.96 | 2.98 | Download | GRGENFMD-IECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVF-HRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWT | |||||||||||||||||||
| 10 | 4gbrA | 0.21 | 0.19 | 0.87 | 4.43 | Download | --------------------DEVWVVGMGIVMSLIVLAIVFGNVLVITAIAKFERLQTVTNYFITSLACADLVMGLAVVPFGAAHILTKTWTFGNFWCEFWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWI | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
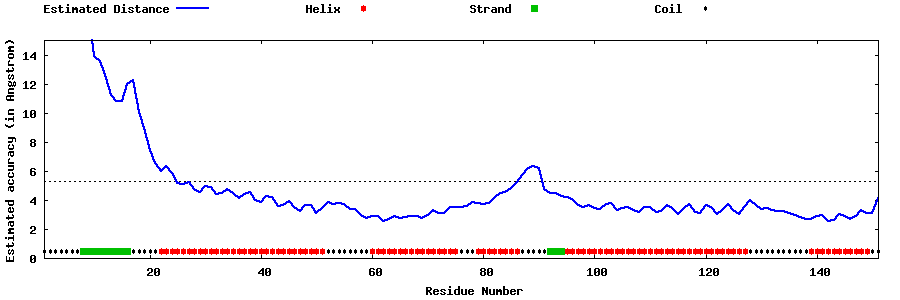 | |||
|
|
|||
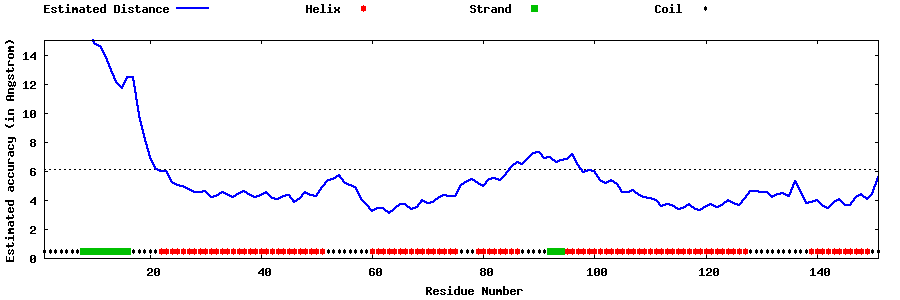 | |||
|
| |||
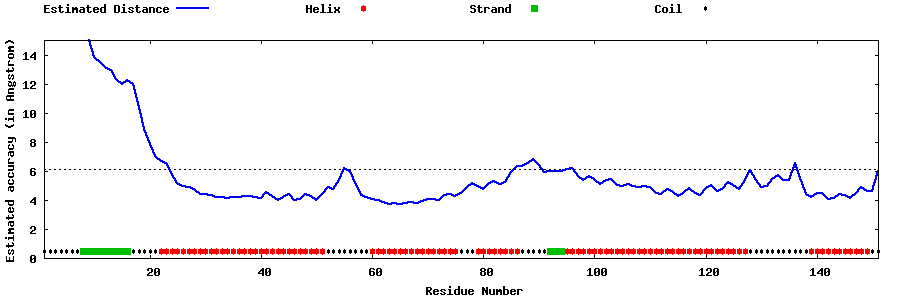 | |||
|
| |||
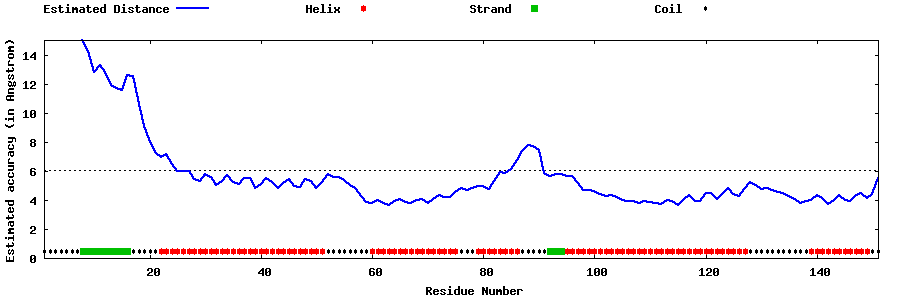 | |||
|
| |||
 | |||
|
| |||