

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MNENNETLTRGFTLMGLFTHNKCSGFFFGVICAVFFMAMIANGVMIFLINIDPHLHTPMYFLLSHLSVIDTLYISTIVPKMLVDYLMGEGTISFIACTAQCFLYMGFMGAEFFLLGLMAYDRYVAICNPLRYPVLISWRVCWMILASSWFGGALDSFLLTPITMSLPFCASHQINHFFCEAPTMLRLACGDKTTYETVMYVCCVAMLLIPFSVVTASYTRILITVHQMTSAEGRKKAFATCSSHMMVVTLFYGAALYTYTLPQSYHTPIKDKVFSAFYTILTPLLNPLIYSLRNRDVMGALKRVVARC | |
| CCCCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHCC | |
| 99988730013488768999445899999999999999998999989998678755738999878999987535007899999853699588589999999999999999999999998650565164545883157869999999999999999999999992348899794788604838888887138619999999999999999899999999999999933877032530766278689979999876886040899999987784887018418764112633146599999999998529 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MNENNETLTRGFTLMGLFTHNKCSGFFFGVICAVFFMAMIANGVMIFLINIDPHLHTPMYFLLSHLSVIDTLYISTIVPKMLVDYLMGEGTISFIACTAQCFLYMGFMGAEFFLLGLMAYDRYVAICNPLRYPVLISWRVCWMILASSWFGGALDSFLLTPITMSLPFCASHQINHFFCEAPTMLRLACGDKTTYETVMYVCCVAMLLIPFSVVTASYTRILITVHQMTSAEGRKKAFATCSSHMMVVTLFYGAALYTYTLPQSYHTPIKDKVFSAFYTILTPLLNPLIYSLRNRDVMGALKRVVARC | |
| 85442302201000000043350210001303332332333333002002001300000011020000000111002002000000175320001002301100120022013000300200000002102000100330000001100310131031000100001003423010000032100300020033102201331333133123203301330010011031451231010000000000011320000000102153355331000021033203300300002042023002300533 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCSSSSSSSCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCCCCCHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCSSSSHHHHHHHCCCCHHHCCCCHHHHHHHHHHHHCC MNENNETLTRGFTLMGLFTHNKCSGFFFGVICAVFFMAMIANGVMIFLINIDPHLHTPMYFLLSHLSVIDTLYISTIVPKMLVDYLMGEGTISFIACTAQCFLYMGFMGAEFFLLGLMAYDRYVAICNPLRYPVLISWRVCWMILASSWFGGALDSFLLTPITMSLPFCASHQINHFFCEAPTMLRLACGDKTTYETVMYVCCVAMLLIPFSVVTASYTRILITVHQMTSAEGRKKAFATCSSHMMVVTLFYGAALYTYTLPQSYHTPIKDKVFSAFYTILTPLLNPLIYSLRNRDVMGALKRVVARC | |||||||||||||||||||||||||
| 1 | 3emlA | 0.17 | 0.23 | 0.89 | 3.53 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFT-FFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| 2 | 5tgzA | 0.16 | 0.22 | 0.92 | 2.29 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEK--------LQSVCSDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFSMLCLLNSTVNPIIYALRSKDLRHAFRSM---- | |||||||||||||||||||
| 3 | 5tgzA | 0.17 | 0.22 | 0.91 | 2.18 | Download | GRGENFMDIEC----FMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKD-SRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLCEKLQSVCSD-IFPHID------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF--- | |||||||||||||||||||
| 4 | 4djh | 0.14 | 0.22 | 0.91 | 1.53 | Download | -------------------SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIRYTKMKTATNIYIFNLALADALVT-TTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKV---REDVDVIECSLQFPD----DDYWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP- | |||||||||||||||||||
| 5 | 4yay | 0.14 | 0.20 | 0.92 | 1.24 | Download | LKTTRNAYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGIDIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL--- | |||||||||||||||||||
| 6 | 3emlA | 0.18 | 0.23 | 0.89 | 3.74 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| 7 | 4iaq | 0.18 | 0.22 | 0.85 | 1.71 | Download | ---------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASE----------CVVN----------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIADARERKATKTLGIILGAFIVCWLPFFIISLVMPIH--L-AIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRF- | |||||||||||||||||||
| 8 | 2z73A | 0.16 | 0.23 | 0.92 | 2.92 | Download | --EYNPSIVREF-----DQVPAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNGPLMTISCFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMSHRRAFIMIIFVWLWSVLWAI---GPIFGWVLCNDYI----------------SRDSTTRSNILCMFILGFFGPILIIFFCYFNIVMAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWVTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWV | |||||||||||||||||||
| 9 | 3emlA | 0.17 | 0.23 | 0.89 | 4.60 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKS-LAIIVGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| 10 | 2ydoA | 0.15 | 0.21 | 0.92 | 5.55 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPLFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH | |||||||||||||||||||
| ||||||||||||||||||||||||||
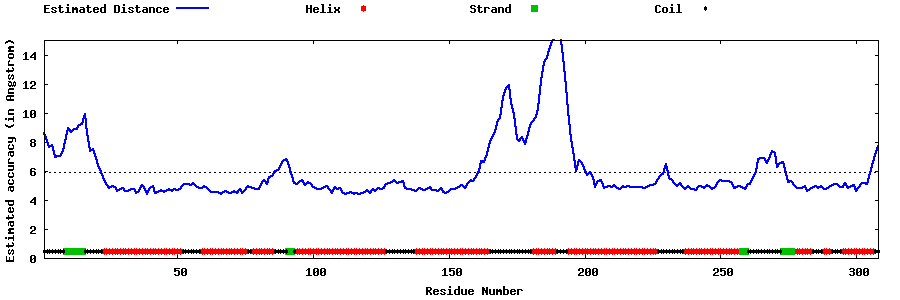
| Generated 3D models | Estimated local accuracy of models | ||
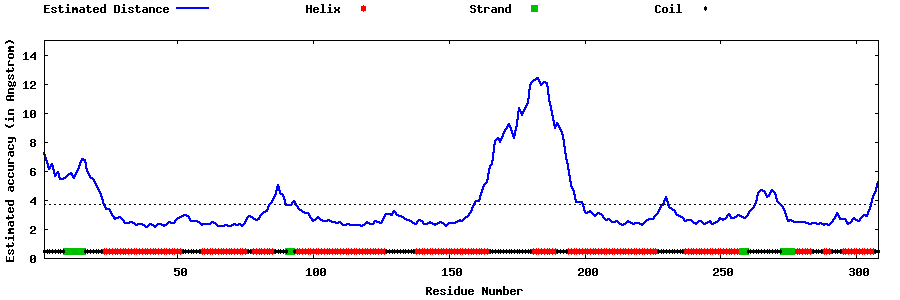 | |||
|
|
|||
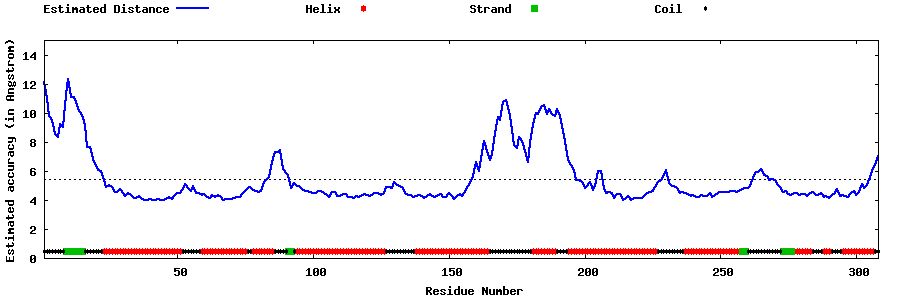 | |||
|
| |||
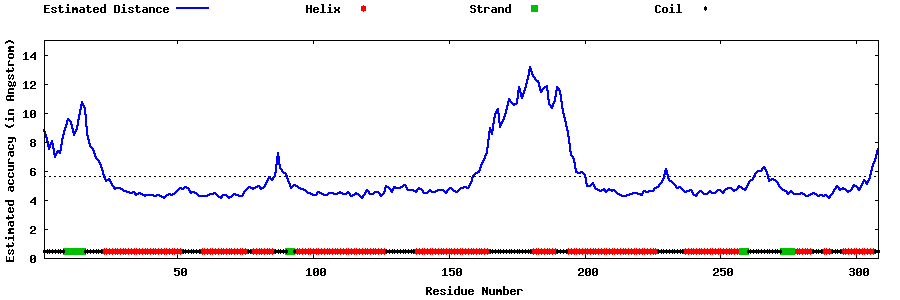 | |||
|
| |||
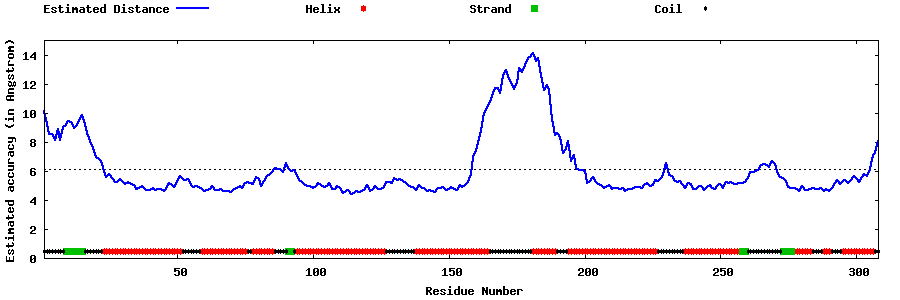 | |||
|
| |||
 | |||
|
| |||