

| 20 40 60 80 100 120 140 160 180 200 | |
| | | | | | | | | | | | |
| MEMTKLFSYIVIKNVYYPQVSFGISANTFLLLFHIFTFAYTHRLKPIDMTISHLPLIHILLLFTQAILVSSDLFESWNIQNNDLKCKIITFLNRVMRGVSICTTCLLSVLQAITISPSTSFLEKFKHISANHTLGFILFSWVLNMFITNNLLLFIVPTPNRIGASLLFVTEHCYVLPMSYTHRSLFFILMVLRDVIFIGLMVLSSGYG | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSSHHHHCCCHHHHHHHHHHHHSSSSCCCCHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCSSCCCCCSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCC | |
| 9701605999999999999999999999999999999983889994999999999999999999648999999964567589875167887607656530657888888722894289638888888567203558999999999986040389866885665267101306825077378888888999897899999998725799 |
| 20 40 60 80 100 120 140 160 180 200 | |
| | | | | | | | | | | | |
| MEMTKLFSYIVIKNVYYPQVSFGISANTFLLLFHIFTFAYTHRLKPIDMTISHLPLIHILLLFTQAILVSSDLFESWNIQNNDLKCKIITFLNRVMRGVSICTTCLLSVLQAITISPSTSFLEKFKHISANHTLGFILFSWVLNMFITNNLLLFIVPTPNRIGASLLFVTEHCYVLPMSYTHRSLFFILMVLRDVIFIGLMVLSSGYG | |
| 6743312230111111331133233233303321010002444331110012101311221031233230020011343333311000000111132232010001012300000013323034023302220113001203211313330023030333333333333321111133333211110311333323112123221328 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSSSSHHHHCCCHHHHHHHHHHHHSSSSCCCCHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCCSSCCCCCSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCCC MEMTKLFSYIVIKNVYYPQVSFGISANTFLLLFHIFTFAYTHRLKPIDMTISHLPLIHILLLFTQAILVSSDLFESWNIQNNDLKCKIITFLNRVMRGVSICTTCLLSVLQAITISPSTSFLEKFKHISANHTLGFILFSWVLNMFITNNLLLFIVPTPNRIGASLLFVTEHCYVLPMSYTHRSLFFILMVLRDVIFIGLMVLSSGYG | |||||||||||||||||||||||||
| 1 | 4grvA | 0.14 | 0.18 | 0.95 | 1.02 | Download | LDNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR-----KSLQTVHYHLGSLALSDLLILLLAMPVELYNFIWVHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAIHPFKAKTL----MSRSRTKKFISAIWLASALLAIPMLFTMGLQNRSADGTHPGGL-VCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVI | |||||||||||||||||||
| 2 | 5x33A | 0.12 | 0.23 | 0.89 | 1.43 | Download | ------FIPLLAMILLSVSMVVGLPGNTFVVW---SILKRMRKRSVTALMVLNLALADLAVLLT--APFFLHFLTWGTWSFGLAGCRLCHYICGVSMYASVLLITAMSLDRSLAVA---SPFLSQKVRTKTAARWLLVGIWGASFLLATPVLAFRKVVKLTNETD------LCLAVYPSDRHKAFHLLFEAF-TGFVVPFLIVVASY- | |||||||||||||||||||
| 3 | 5t1aA | 0.16 | 0.21 | 0.90 | 1.28 | Download | ---VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILINC--KKLK-CLTDIYLLNLAISDLLFLIT--LPLWAHSAAN-EWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIV-HAVFALKART--VTFGVVTSVITWLVAVFASVPGIIFT--KQKEDSVYV------CGPYFPRGWNNFHTIMRNILGLVLPLLIMVICYSGI | |||||||||||||||||||
| 4 | 5t1a | 0.16 | 0.21 | 0.90 | 1.55 | Download | ---VKQIGAQLLPPLYSLVFIFGFVGNMLVVLILIN---CKKLKCLTDIYLLNLAISDLLFLIT--LPLWAHSA-ANEWVFGNAMCKLFTGLYHIGYFGGIFFIILLTIDRYLAIVHAVFAL---KARTVTFGVVTSVITWLVAVFASVPGIIFTKQKE--------DSVYVCGPYFPRGWNNFHTIMRNILGLVLPLLIMVICYSGI | |||||||||||||||||||
| 5 | 5x33 | 0.13 | 0.17 | 0.92 | 1.22 | Download | ---SNTFIPLLAMILLSVSMVVGLPGNTFVVWSILK---RMRKRSVTALMVLNLALADLAVLL--TAPFFLHFLTWGTWSFGLAGCRLCHYICGVSMYASVLLITAMSLDRSLAVASPF--LS-QKVRTKTAARWLLVGIWGASFLLATPVLAFRKVVKL------TNETDLCLAVYPSDRHKAFHLLEAFTGFVVPFLIVVASYADI | |||||||||||||||||||
| 6 | 4grvA | 0.14 | 0.18 | 0.95 | 1.15 | Download | --NTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLAR----KSLQSTVHYHLGSLALSDLLILLLAMPVELYNFIWHHPWAFGDAGCRGYYFLRDACTYATALNVASLSVARYLAICHPFKAKTL---MSRSRTKKFISAIWLASALLAIPMLFTMGL-QNRSADGTHPGGLVCTPIVDTATVKVVIQVNTFMSFLFPMLVISILNTVI | |||||||||||||||||||
| 7 | 4xnv | 0.15 | 0.22 | 0.88 | 1.67 | Download | -----GFQFYYLPAVYILVFIIGFLGNSVAIWMF---VFHMKPWSGISVYMFNLALADFLYVLT--LPALIFYFNKTDWIFGDAMCKLQRFIFHVNLYGSILFLTCISAHRYSGVVYP-KSLGRLKK---KNAICISVLVWLIVVVAISPILFYSGTGVRK------NKTITCYDTTSDEYLYFIYSMTTVAMFCVPLVLILG----- | |||||||||||||||||||
| 8 | 4dklA | 0.18 | 0.24 | 0.88 | 1.18 | Download | -----MVTAITIMALYSIVCVVGLFGNFLVMYV---IVRYTKMKTATNIYIFNLALADALATST--LPFQSVNYLMGTWPFGNILCKIVISIDYYNMFTSIFTLCTMSVDRYIAVCHPVKALDFR---TPRNAKIVNVCNWILSSAIGLPVMFMIDCTLTFSH-------------PTWYWENLLKICVFIFAFIMPVLIITVCYGLM | |||||||||||||||||||
| 9 | 5unfA | 0.16 | 0.16 | 0.71 | 1.14 | Download | -------HLDAIPILYYIIFVIGFLVNIVVVTLFCCQ----GPKKVSSIYIFNLAVADLLLLATLPLWATYYSYRY-DWLFGPVMCKVFGSFLTLNMFASIFFITCMSVDRYQSVYP-------FPWQA----SYIVPLVWCMACLSSLPTFYFRDVRTIEYLGVNA-----CIMA-------------------------------- | |||||||||||||||||||
| 10 | 6b73A | 0.16 | 0.19 | 0.90 | 1.75 | Download | LGSISPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----YTKMKTTNIYIFNLALADALV--TTTMPFQSTVYLMNSWPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC--HPVKAL-DFRTPLKAKIINICIWLLSSSVGISAIVLGG--------TKVRDVIECSLQFPDDLFMKICVFIFAF--VIPVLIIIVC--YT | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
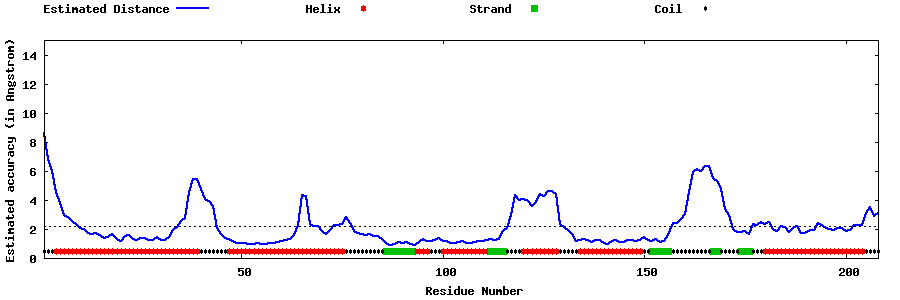 | |||
|
|
|||
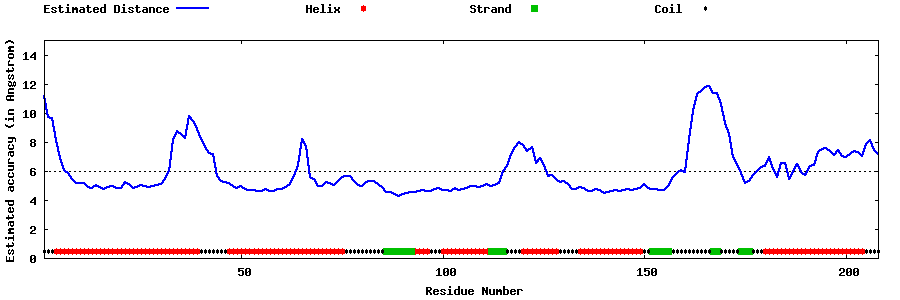 | |||
|
| |||
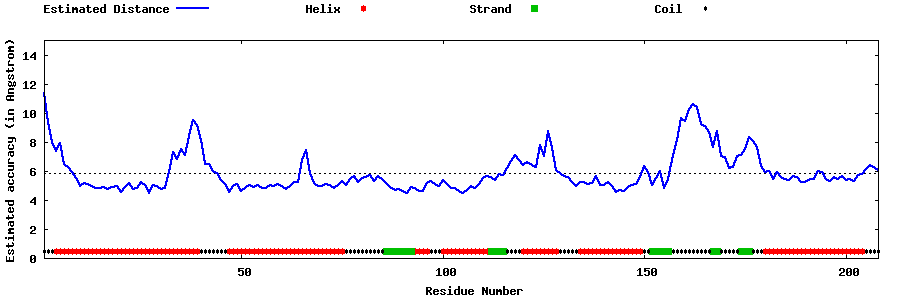 | |||
|
| |||
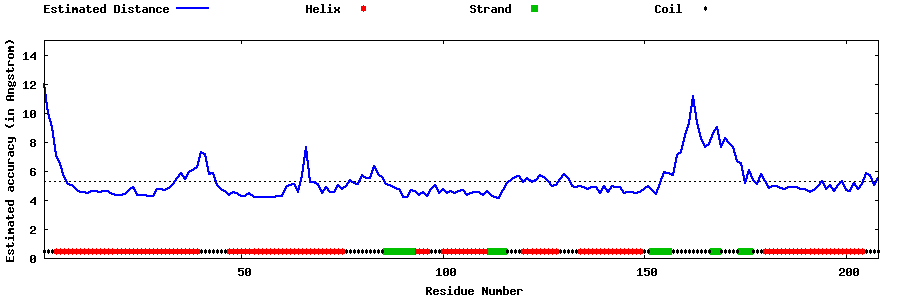 | |||
|
| |||
 | |||
|
| |||