

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MVTEFIFLGLSDSQELQTFLFMLFFVFYGGIVFGNLLIVITVVSDSHLHSPMYFLLANLSLIDLSLSSVTAPKMITDFFSQRKVISFKGCLVQIFLLHFFGGSEMVILIAMGFDRYIAICKPLHYTTIMCGNACVGIMAVAWGIGFLHSVSQLAFAVHLPFCGPNEVDSFYCDLPRVIKLACTDTYRLDIMVIANSGVLTVCSFVLLIISYTIILMTIQHCPLDKSSKALSTLTAHITVVLLFFGPCVFIYAWPFPIKSLDKFLAVFYSVITPLLNPIIYTLRNKDMKTAIRRLRKWDAHSSVKF | |
| CSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCHHHHHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHHCCCCCCCCC | |
| 91269853699883268999999999999999878874541321799999789998749988676432271999999735997786899999999999999999999999986518873623146021687599999999999999999999999610899898836686318188888940331455645766666999999999999999999999864632130999998879992336630532488688999961355578886334400037443562999999999997116664479 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MVTEFIFLGLSDSQELQTFLFMLFFVFYGGIVFGNLLIVITVVSDSHLHSPMYFLLANLSLIDLSLSSVTAPKMITDFFSQRKVISFKGCLVQIFLLHFFGGSEMVILIAMGFDRYIAICKPLHYTTIMCGNACVGIMAVAWGIGFLHSVSQLAFAVHLPFCGPNEVDSFYCDLPRVIKLACTDTYRLDIMVIANSGVLTVCSFVLLIISYTIILMTIQHCPLDKSSKALSTLTAHITVVLLFFGPCVFIYAWPFPIKSLDKFLAVFYSVITPLLNPIIYTLRNKDMKTAIRRLRKWDAHSSVKF | |
| 63100000000424400100013133323323332330000020043030000100230012001100000020000002532100050000000110331131020002003100000022110200013310000012013102201300131023030000122000100221002000020230103002301310231033133112100200132146122200100300210030232101000000333233012100310231033113000020540230033014431435753 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CSSSSSSCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHSSSSSCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHCCSSSSSCCCCCCCCCHHHHHHHHHHHHHHCCCHHHCCCCHHHHHHHHHHHHCCCCCCCCC MVTEFIFLGLSDSQELQTFLFMLFFVFYGGIVFGNLLIVITVVSDSHLHSPMYFLLANLSLIDLSLSSVTAPKMITDFFSQRKVISFKGCLVQIFLLHFFGGSEMVILIAMGFDRYIAICKPLHYTTIMCGNACVGIMAVAWGIGFLHSVSQLAFAVHLPFCGPNEVDSFYCDLPRVIKLACTDTYRLDIMVIANSGVLTVCSFVLLIISYTIILMTIQHCPLDKSSKALSTLTAHITVVLLFFGPCVFIYAWPFPIKSLDKFLAVFYSVITPLLNPIIYTLRNKDMKTAIRRLRKWDAHSSVKF | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.24 | 0.90 | 3.42 | Download | -------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS---GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVPMNYMVYF-----------NFFACVLVPLLLMLGVYLRIFLAARRQVHAAKSLAIIVGLFALCWLPLH-IINCFTFFCPDCAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.28 | 0.87 | 2.17 | Download | ------------NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSM----------- | |||||||||||||||||||
| 3 | 4iaqA | 0.20 | 0.24 | 0.82 | 2.10 | Download | -----YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISILYTVYSTVGAF---------------------------------------YFPTLLLIALYGRIYVEARSRIIQKAARERKATKTLGIILGAFIVCWLPFFIISLVMPLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 4 | 4ib4 | 0.19 | 0.23 | 0.90 | 1.54 | Download | ----------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMEAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIE-T-----NPNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR----- | |||||||||||||||||||
| 5 | 4yay | 0.17 | 0.22 | 0.90 | 1.20 | Download | YIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK-----KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDVLIVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL---------- | |||||||||||||||||||
| 6 | 3emlA | 0.21 | 0.24 | 0.90 | 3.51 | Download | --------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQVHAAKSLAIIVGLFALCWLPLHII-NCFTFFCPDCSHAWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ--- | |||||||||||||||||||
| 7 | 4iaq | 0.20 | 0.26 | 0.86 | 1.71 | Download | -------------LPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFFW-RQASE----------CVVNT----------DHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH-LAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK------- | |||||||||||||||||||
| 8 | 4buoA | 0.17 | 0.22 | 0.90 | 2.81 | Download | D------LDVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPGVCTPI--------VDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQAVVIAFVVCWLPYHVRRLMFCYI-------SDEQWYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL---------- | |||||||||||||||||||
| 9 | 5tgzA | 0.22 | 0.28 | 0.89 | 4.45 | Download | DIECFMVL----NPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFH-RKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG---------NCEK---------LQSVCSDIFHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARELAKTLVLILVVLIICWGPLLAIMVYDVFGKMTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF---------- | |||||||||||||||||||
| 10 | 2ydoA | 0.18 | 0.23 | 0.94 | 5.58 | Download | ----------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQESTLQKEVHAAKSLAIIVGLFALCCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQQEP | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
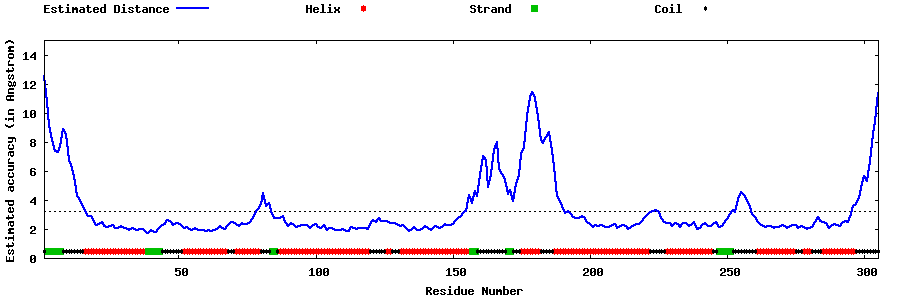 | |||
|
|
|||
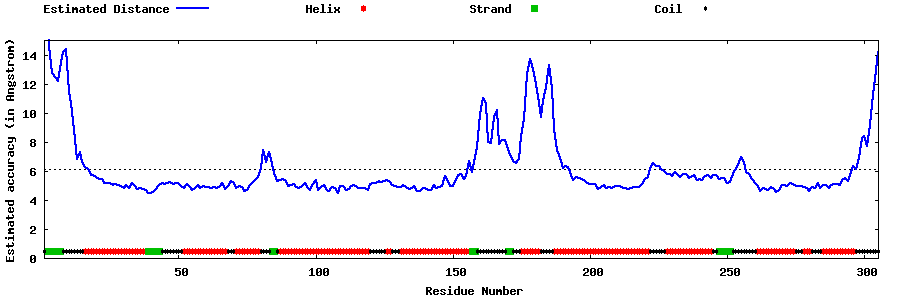 | |||
|
| |||
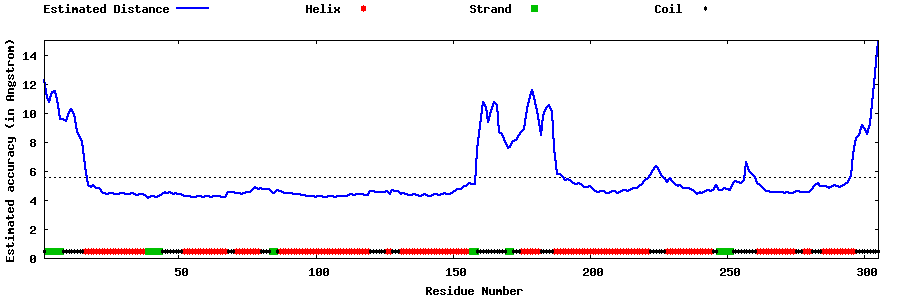 | |||
|
| |||
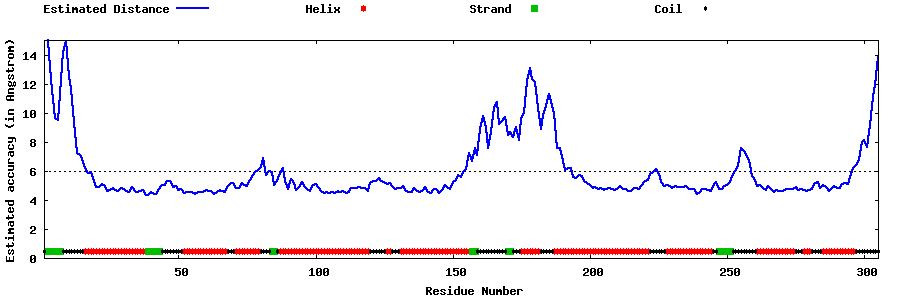 | |||
|
| |||
 | |||
|
| |||