

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MRQINQTQVTEFLLLGLSDGPHTEQLLFIVLLGVYLVTVLGNLLLISLVHVDSQLHTPMYFFLCNLSLADLCFSTNIVPQALVHLLSRKKVIAFTLCAARLLFFLIFGCTQCALLAVMSYDRYVAICNPLRYPNIMTWKVCVQLATGSWTSGILVSVVDTTFILRLPYRGSNSIAHFFCEAPALLILASTDTHASEMAIFLMGVVILLIPVFLILVSYGRIIVTVVKMKSTVGSLKAFSTCGSHLMVVILFYGSAIITYMTPKSSKQQEKSVSVFYAIVTPMLNPLIYSLRNKDVKAALRKVATRNFP | |
| CCCCCCCSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCC | |
| 99988733556667569998517999999999999999998899999997588756758999878999877776148899999846799678589999999999999999999999998650665165545882258879999999999999999999999992408899895788614828999886247509999999999999999899999999999999813866265651420658789989999864762361789987678289865405333213563315659999999999707294 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MRQINQTQVTEFLLLGLSDGPHTEQLLFIVLLGVYLVTVLGNLLLISLVHVDSQLHTPMYFFLCNLSLADLCFSTNIVPQALVHLLSRKKVIAFTLCAARLLFFLIFGCTQCALLAVMSYDRYVAICNPLRYPNIMTWKVCVQLATGSWTSGILVSVVDTTFILRLPYRGSNSIAHFFCEAPALLILASTDTHASEMAIFLMGVVILLIPVFLILVSYGRIIVTVVKMKSTVGSLKAFSTCGSHLMVVILFYGSAIITYMTPKSSKQQEKSVSVFYAIVTPMLNPLIYSLRNKDVKAALRKVATRNFP | |
| 87462302001000000043240111001302331331333332002001002300000011020000000101003203001001164320100002201210131033003000100200000002102000200330000001100110031031000000001003624010000032100200021032001202331333133123103201320010001032461231011000000000001120000000104235533000002103320331030010204201300230044628 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCSSSSSSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHCCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCCHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHCHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCSSSSSHHCHHHCCCCHHHCCCCHHHHHHHHHHHHCCCC MRQINQTQVTEFLLLGLSDGPHTEQLLFIVLLGVYLVTVLGNLLLISLVHVDSQLHTPMYFFLCNLSLADLCFSTNIVPQALVHLLSRKKVIAFTLCAARLLFFLIFGCTQCALLAVMSYDRYVAICNPLRYPNIMTWKVCVQLATGSWTSGILVSVVDTTFILRLPYRGSNSIAHFFCEAPALLILASTDTHASEMAIFLMGVVILLIPVFLILVSYGRIIVTVVKMKSTVGSLKAFSTCGSHLMVVILFYGSAIITYMTPKSSKQQEKSVSVFYAIVTPMLNPLIYSLRNKDVKAALRKVATRNFP | |||||||||||||||||||||||||
| 1 | 3emlA | 0.19 | 0.22 | 0.89 | 3.49 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITISTG--FCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 2 | 5tgzA | 0.22 | 0.28 | 0.91 | 2.20 | Download | -RGENFMDIECF----MVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVL--------PLLGWNCEKL---------QSVCSDIFPHDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARMELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVMLCLLNSTVNPIIYALRSKDLRHAFRSM------ | |||||||||||||||||||
| 3 | 4iaqA | 0.22 | 0.25 | 0.87 | 2.29 | Download | ------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGR----WVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-------PPFFWRQAS--------------ECVVNTDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKRERKATKTLGIILGAFIVCWLPFFIISLVM-PIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK-- | |||||||||||||||||||
| 4 | 4ib4 | 0.21 | 0.24 | 0.90 | 1.52 | Download | -----------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIK--GIE----TNPNNITCVLTK--------E-RFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLNEQRASKVLGIVFFLFLLMWCPFFITNITVLCNQQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR | |||||||||||||||||||
| 5 | 4yay | 0.20 | 0.23 | 0.91 | 1.24 | Download | KTTRN-AYIQKYLILNSSDCNYIFVMIPTLYSIIFVVGIFGNSLVVIVIYFYMKLKTVASVFLLNLALADLCFLLTLPLWAVYTAMEYRWPFGNYLCKIASASVSFNLYASVFLLTCLSIDRYLAIVHPMKSRLRRTMLVAKVTCIIIWLLAGLASLPAIIHRNVFF-------IITVCAFHYE--------TLPIGLGLTKNILGFLFPFLIILTSYTLIWKALK------KNDDIFKIIMAIVLFFFFSWIPHQIFTFLDLGDVDTAMPITICIAYFNNCLNPLFYGFLGKKFKRYFLQLL----- | |||||||||||||||||||
| 6 | 3emlA | 0.20 | 0.22 | 0.89 | 3.78 | Download | ---------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| 7 | 3uon | 0.18 | 0.27 | 0.87 | 1.72 | Download | ------------------------VFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF------S---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM---- | |||||||||||||||||||
| 8 | 4ea3A | 0.21 | 0.20 | 0.83 | 3.07 | Download | -------------------PLGLKVTIVGLYLAVCVGGLLGNCLVMYVILRHTKMKTATNIYIFNLALADTLVLL-TLPFQGTDILLGFWPFGNALCKTVIAIDYYNMFTSTFTLTAMSVDRYVAICHPTSSKAQAVNVAIWALASVVGVPVAI--------MGSAQVEDEEIIPT-------------PQDYWGPVFAICIFLFSFIVPVLVISVCYSLMIRRLRGVRLLSGSVAVFVGCWTPVQVFVLAQGLG----VQPSSTAVAILRFCTALGYVNSCLNPILYAFLDENFKACFR-------- | |||||||||||||||||||
| 9 | 3emlA | 0.20 | 0.22 | 0.88 | 4.62 | Download | --------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIS--TGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAI-IVGLFALCWLPLHIINCFTFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRRQ-- | |||||||||||||||||||
| 10 | 2ydoA | 0.17 | 0.24 | 0.92 | 5.55 | Download | -----------------------SSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSAAAADILVGVLAIPFAIA--ISTGFCAACHGCLFIACFVLVLTASSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGQPKEGKAHSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLKQMESTLQKEVHAAKSLAIIVGLFALCWLPFFCPDCSHAPLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVL | |||||||||||||||||||
| ||||||||||||||||||||||||||
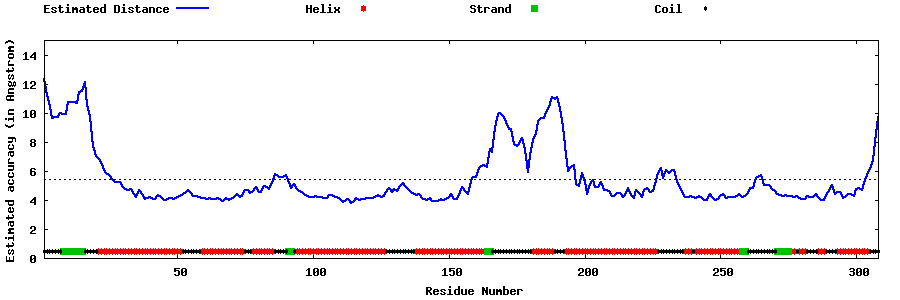
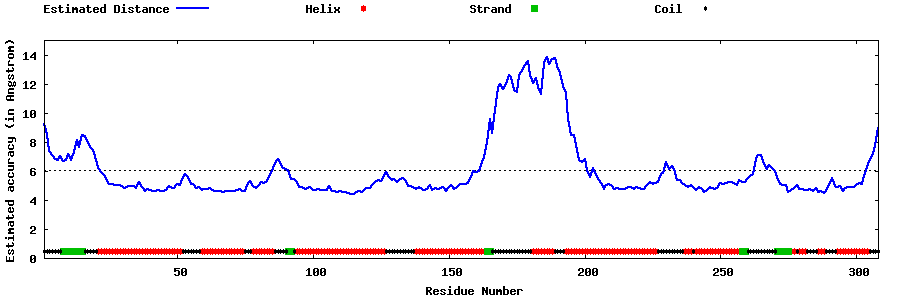
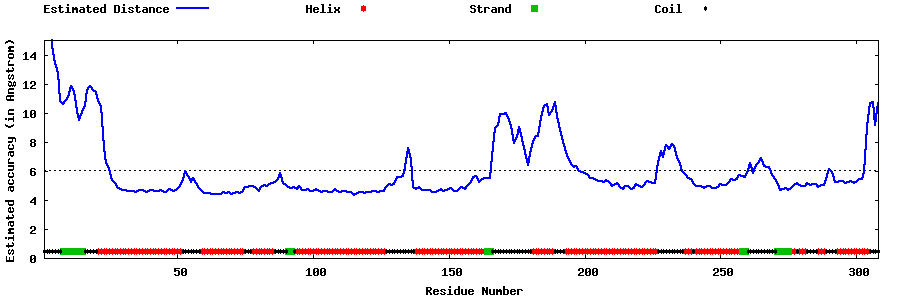
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||