

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MESGLLRPAPVSEVIVLHYNYTGKLRGARYQPGAGLRADAVVCLAVCAFIVLENLAVLLVLGRHPRFHAPMFLLLGSLTLSDLLAGAAYAANILLSGPLTLKLSPALWFAREGGVFVALTASVLSLLAIALERSLTMARRGPAPVSSRGRTLAMAAAAWGVSLLLGLLPALGWNCLGRLDACSTVLPLYAKAYVLFCVLAFVGILAAICALYARIYCQVRANARRLPARPGTAGTTSTRARRKPRSLALLRTLSVVLLAFVACWGPLFLLLLLDVACPARTCPVLLQADPFLGLAMANSLLNPIIYTLTNRDLRHALLRLVCCGRHSCGRDPSGSQQSASAAEASGGLRRCLPPGLDGSFSGSERSSPQRDGLDTSGSTGSPGAPTAARTLVSEPAAD | |
| CCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC | |
| 98666899987642122121579878988898339999999999999999998998414312268999589999999999999999999999999763531516701888711678879999999999999859880587769860387889988999999999999999862877799831478404786055763589999999999999999999999999997436776664201356688999999999999999999999999999999779877873479999999999999996999993799999999999578888999898887777651105898333379877788789888997778998988778999988777666799998 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | |
| | | | | | | | | | | | | | | | | | | | | |
| MESGLLRPAPVSEVIVLHYNYTGKLRGARYQPGAGLRADAVVCLAVCAFIVLENLAVLLVLGRHPRFHAPMFLLLGSLTLSDLLAGAAYAANILLSGPLTLKLSPALWFAREGGVFVALTASVLSLLAIALERSLTMARRGPAPVSSRGRTLAMAAAAWGVSLLLGLLPALGWNCLGRLDACSTVLPLYAKAYVLFCVLAFVGILAAICALYARIYCQVRANARRLPARPGTAGTTSTRARRKPRSLALLRTLSVVLLAFVACWGPLFLLLLLDVACPARTCPVLLQADPFLGLAMANSLLNPIIYTLTNRDLRHALLRLVCCGRHSCGRDPSGSQQSASAAEASGGLRRCLPPGLDGSFSGSERSSPQRDGLDTSGSTGSPGAPTAARTLVSEPAAD | |
| 74442255232331010010211434435322211000000000100210331330000000103300000000000001000101212111000103120110000000100000000000120000001000000010121431332000000000011012102100000012343610000001332000000022133112000200010000003003303434544444443343343011001000000200100010000000000003231000001000001000100021000100104400300130000022234446633544433354444444433443545344363444454444353444443434434433454668 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 340 360 380 | | | | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSSSSCCCCCCSSSSSSHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHCCHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCSSCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCCC MESGLLRPAPVSEVIVLHYNYTGKLRGARYQPGAGLRADAVVCLAVCAFIVLENLAVLLVLGRHPRFHAPMFLLLGSLTLSDLLAGAAYAANILLSGPLTLKLSPALWFAREGGVFVALTASVLSLLAIALERSLTMARRGPAPVSSRGRTLAMAAAAWGVSLLLGLLPALGWNCLGRLDACSTVLPLYAKAYVLFCVLAFVGILAAICALYARIYCQVRANARRLPARPGTAGTTSTRARRKPRSLALLRTLSVVLLAFVACWGPLFLLLLLDVACPARTCPVLLQADPFLGLAMANSLLNPIIYTLTNRDLRHALLRLVCCGRHSCGRDPSGSQQSASAAEASGGLRRCLPPGLDGSFSGSERSSPQRDGLDTSGSTGSPGAPTAARTLVSEPAAD | |||||||||||||||||||||||||
| 1 | 5tjvA | 0.27 | 0.22 | 0.72 | 2.73 | Download | ------------------ENFMDIECFMVLNP-SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKA-------------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKL-IKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------------------- | |||||||||||||||||||
| 2 | 2ziy | 0.16 | 0.19 | 0.89 | 1.51 | Download | -DLRDNETWWYNPSIIVHPHWRE----FDQVPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKTKSLQTPANMFIINLAFSDFTFSLVNFPLMTICFLKKWIFGFAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGRPMAASKKMHRRAFIMIIFVWLWSVLWAIGPIFGWGAYTVLCNCSFDYISTTRSNILCMILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNALRKAQAGANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPLEWV--TPYAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDK-E--TEDDKDAE-------T-----------------EIPAGESSDAAP--------SADAAQMK | |||||||||||||||||||
| 3 | 4z34A | 0.31 | 0.28 | 0.78 | 3.76 | Download | -----EPQCFYNESIAFFYNRSGKHLATEWNT--VSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETKTTRNAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDVLAYEKF--FLLLAEFNSAMNPIIYSYRDKEMSATFRQIL----------------------------------------------------------------------------- | |||||||||||||||||||
| 4 | 5tgzA | 0.27 | 0.22 | 0.73 | 3.32 | Download | --------------GGRGENFMDIECFMVLNPS-QQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHI--DKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHA--------------PDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK-LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------------------- | |||||||||||||||||||
| 5 | 4z34 | 0.31 | 0.28 | 0.78 | 1.57 | Download | ----NEPQCFYNESIAFFYNRSGKHLATEW--NTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFR-MQLHTRMNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETLKAMKDFIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQC--D-VLAEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------------------------------------------- | |||||||||||||||||||
| 6 | 4ib4 | 0.22 | 0.20 | 0.75 | 1.18 | Download | ----------------------------EEQ-GNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANYNSRATAFIKITVVWLISIGIAIPPIKGIETNPNNITCVLTKERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADLEDNWETLAYIQKYLQTISNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQTLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR------------------------------------------------------------------------ | |||||||||||||||||||
| 7 | 5tjvA | 0.27 | 0.22 | 0.72 | 2.92 | Download | -------------------NFMDIECFMV-LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAG-------------KRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNK-LIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------------------- | |||||||||||||||||||
| 8 | 4z34 | 0.32 | 0.28 | 0.77 | 1.80 | Download | ----------YNESIAFFYNRSGKHLATEW--NTVSKLVMGLGITVCIFIMLANLLVMVAIYVNRRFHFPIYYLMANLAAADFFAGLAYFYLMFNTGPNTRRLTVSTWLLRQGLIDTSLTASVANLLAIAIERHITVFRMQLHTRMSNRRVVVVIVVIWTMAIVMGAIPSVGWNCICDIENCSNMAPLYSDSYLVFWAIFNLVTFVVMVVLYAHIFGYVADLEDNWETLNAMKAYIQKYLRNRDTMMSLLKTVVIVLGAFIICWTPGLVLLLLDVCCPQCDV--LAYEKFFLLLAEFNSAMNPIIYSYRDKEMSATFRQILG---------------------------------------------------------------------------- | |||||||||||||||||||
| 9 | 1u19A | 0.18 | 0.20 | 0.83 | 4.75 | Download | -----MNGTEGPNFYVPFSNKTGVVRSPFEAPQWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYGMQCSCGIDYYTPHSFVIYMFVVHFIIPLIVIFFCYGQLVFT----------VKEAAAQQQESATTQKAEKEVTRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAF-FAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPLGDDEASTTVSKTESQVAPA--------------------------------------------------- | |||||||||||||||||||
| 10 | 5tjvA | 0.27 | 0.22 | 0.72 | 2.96 | Download | ------------------ENFMDIECFMVLNP-SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKA-------------GKRAMSFSDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKL-IKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF----------------------------------------------------------------------------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Generated 3D models | Estimated local accuracy of models | ||
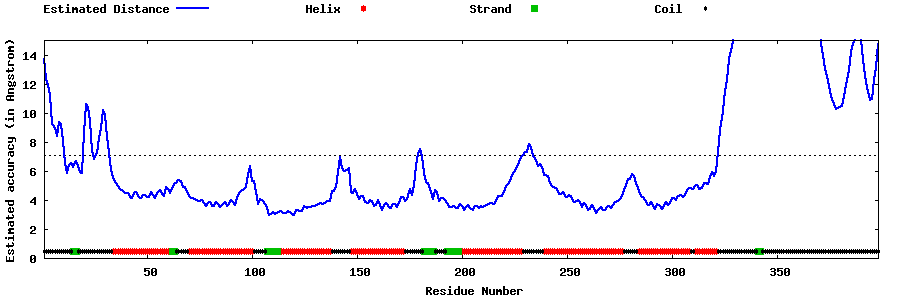 | |||
|
|
|||
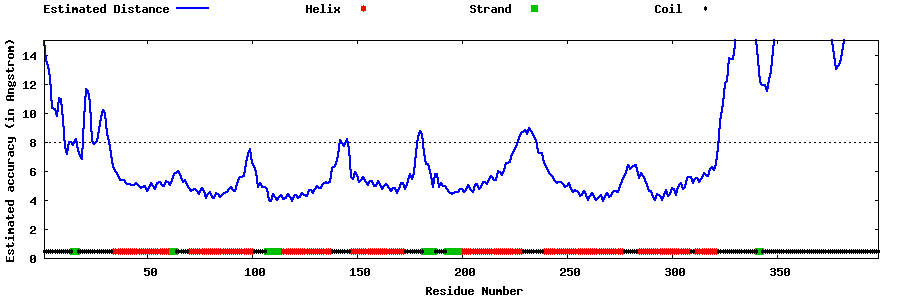 | |||
|
| |||
 | |||
|
| |||
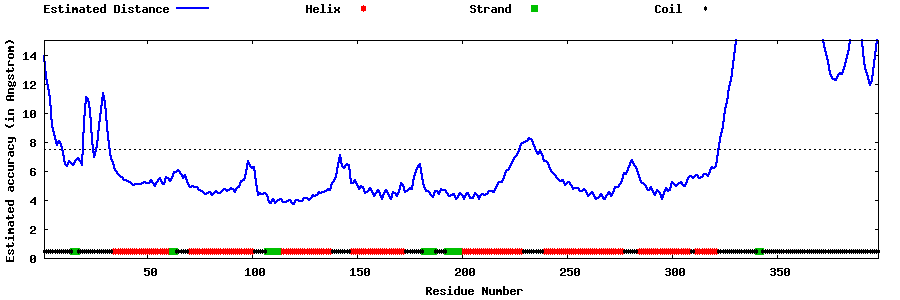 | |||
|
| |||
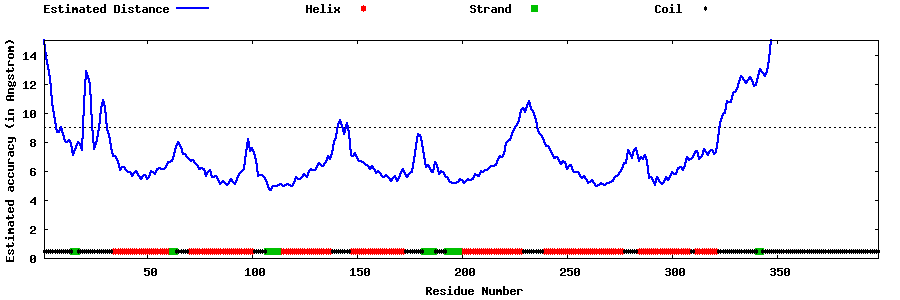 | |||
|
| |||