

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MSDSNLSDNHLPDTFFLTGIPGLEAAHFWIAIPFCAMYLVALVGNAALILVIAMDNALHAPMYLFLCLLSLTDLALSSTTVPKMLAILWLHAGEISFGGCLAQMFCVHSIYALESSILLAMAFDRYVAICNPLRYTTILNHAVIGRIGFVGLFRSVAIVSPFIFLLRRLPYCGHRVMTHTYCEHMGIARLACANITVNIVYGLTVALLAMGLDSILIAISYGFILHAVFHLPSHDAQHKALSTCGSHIGIILVFYIPAFFSFLTHRFGHHEVPKHVHIFLANLYVLVPPVLNPILYGARTKEIRSRLLKLLHLGKTSI | |
| CCCCCCCCCCCCCHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCCC | |
| 999789999786121157998986689999999999999999999999998857887542289999999999998998559999999956998578888999999999999999999999982306621566556312888999999999999999999989999963899998945861214576688825784587899999999999998999999999999998168887688898541599999999999989998102104799996089999999997288764681221657999999999962476889 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | |
| | | | | | | | | | | | | | | | | |
| MSDSNLSDNHLPDTFFLTGIPGLEAAHFWIAIPFCAMYLVALVGNAALILVIAMDNALHAPMYLFLCLLSLTDLALSSTTVPKMLAILWLHAGEISFGGCLAQMFCVHSIYALESSILLAMAFDRYVAICNPLRYTTILNHAVIGRIGFVGLFRSVAIVSPFIFLLRRLPYCGHRVMTHTYCEHMGIARLACANITVNIVYGLTVALLAMGLDSILIAISYGFILHAVFHLPSHDAQHKALSTCGSHIGIILVFYIPAFFSFLTHRFGHHEVPKHVHIFLANLYVLVPPVLNPILYGARTKEIRSRLLKLLHLGKTSI | |
| 444314243331110200000121320200011022012202321210000021151002000200010010000002000020000000204403040000001200310130100011002000000020010000003310010000003213011202100021032045200000001021001000230300111002003312332231033011200200020024601320120010010001003231321010000033300210000002310030033003000020340041003002445665 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCCCCCCCCCCCHHSSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCHHHHHHHHHHHHCCCCCCCSSSSCCHHHHHHHHHHHHCCCCCCC MSDSNLSDNHLPDTFFLTGIPGLEAAHFWIAIPFCAMYLVALVGNAALILVIAMDNALHAPMYLFLCLLSLTDLALSSTTVPKMLAILWLHAGEISFGGCLAQMFCVHSIYALESSILLAMAFDRYVAICNPLRYTTILNHAVIGRIGFVGLFRSVAIVSPFIFLLRRLPYCGHRVMTHTYCEHMGIARLACANITVNIVYGLTVALLAMGLDSILIAISYGFILHAVFHLPSHDAQHKALSTCGSHIGIILVFYIPAFFSFLTHRFGHHEVPKHVHIFLANLYVLVPPVLNPILYGARTKEIRSRLLKLLHLGKTSI | |||||||||||||||||||||||||
| 1 | 3emlA | 0.20 | 0.23 | 0.86 | 2.42 | Download | ----------------------IM-GSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACL-----------FEDVVPMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSH---V | |||||||||||||||||||
| 2 | 5tgzA | 0.17 | 0.25 | 0.85 | 2.13 | Download | ------------------------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDF-HVFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLL-------GWNCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKAHSHAPDQARIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 3 | 5tgzA | 0.16 | 0.25 | 0.86 | 1.80 | Download | GENFMDIECFMNP-----------SQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFH-VFHRKDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG----WNCEKLIFPHID--------------------KTYLMFWIGVVSVLLLFIVYAYMYILWKAHSQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 4 | 4ib4 | 0.17 | 0.21 | 0.88 | 1.52 | Download | --------------------EEQGNKLHWAALLILMVIIPTIGGNTLVILAVSLEKKLQYATNYFLMSLAVADLLVGLFVMPIALLTIMFAMWPLPLVLCPAWLFLDVLFSTASIWHLCAISVDRYIAIKKPIQANQYNSRATAFIKITVVWLISIGIAIPVPIKGIE-TN-----PNNITCVLTK---------ERFGDFMLFGSLAAFFTPLAIMIVTYFLTIHALQKKAADSNEQRASKVLGIVFFLFLLMWCPFFITNITLVLCDSCNQLQMLLEIFVWIGYVSSGVNPLVYTLFNKTFRDAFGRYITCNYR-- | |||||||||||||||||||
| 5 | 3uonA | 0.18 | 0.17 | 0.86 | 1.12 | Download | -----------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFI--VGVRTVEDGECYIQF---------FSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSR----REKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTV-WTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------ | |||||||||||||||||||
| 6 | 3emlA | 0.19 | 0.23 | 0.87 | 2.66 | Download | ------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVV-----------PMNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCFTFFC----PDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 4iaq | 0.19 | 0.21 | 0.83 | 1.69 | Download | ------------------------PWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISLPPFF-WRQASECVVN--------------------TDHILYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIAAARERKATKTLGIILGAFIVCWLPFFIISLVMPIH----L-AIFD-FFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK---- | |||||||||||||||||||
| 8 | 4buoA | 0.17 | 0.20 | 0.90 | 3.12 | Download | -------NSDL-------DVNTDIYSKVLVTAIYLALFVVGTVGNSVTLFTLARKKSLQSTVDYYLGSLALSDLLILLLAMPVELYNFIWVHHPWAFAGCRGYYFLRDACTYATALNVVSLSVELYLAICHPFKAKTLMSRSRTKKFISAIWLASALLAIPMLFTMGLQNLSGDGTHPG------------GVDTATLKVVIQVNTFMSFLFPMLVASILNTVIANKLTVMVHQPGRVQALRRGVLVLRAVVIAFVVCWLPYHVRRLMFCYISDHYFYMLTNALVYVSAAINPILYNLVSANFRQVFLSTL------- | |||||||||||||||||||
| 9 | 5tgzA | 0.17 | 0.25 | 0.90 | 3.16 | Download | --GGRGENFMDIECFMVLN----PSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRK-DSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLG--------NCEKLQSVC---------SDIFPHIDKTYLMFWIGVVSVLLLFIVYAYMYILWKA-APDQARMDIELAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------- | |||||||||||||||||||
| 10 | 4gpoA | 0.19 | 0.22 | 0.87 | 4.59 | Download | ----------------------LQQWEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASIETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMMHWWRDEDPQALKCYQDPG--------CCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKEQVMLMREHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRDLVP-DWLFVAFNWLGYANSAMNPIIYC-RSPDFRKAFKRL-------- | |||||||||||||||||||
| ||||||||||||||||||||||||||
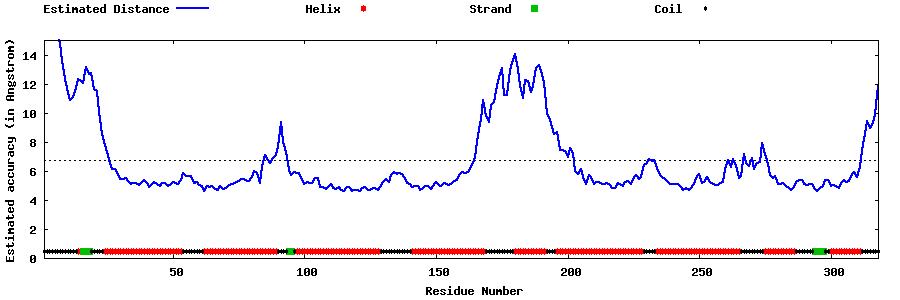
| Generated 3D models | Estimated local accuracy of models | ||
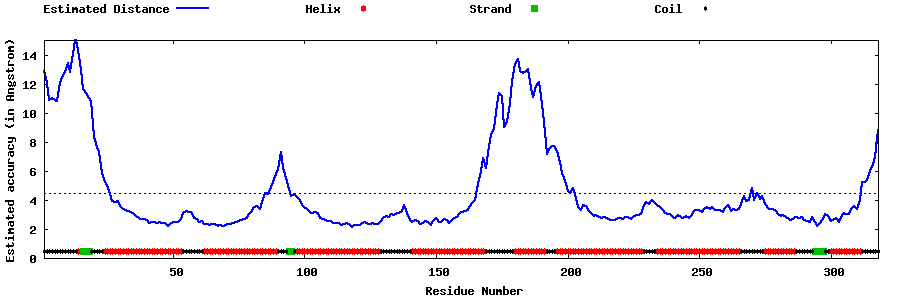 | |||
|
|
|||
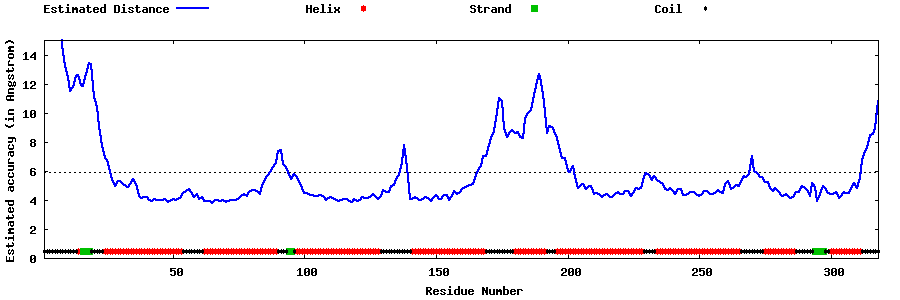 | |||
|
| |||
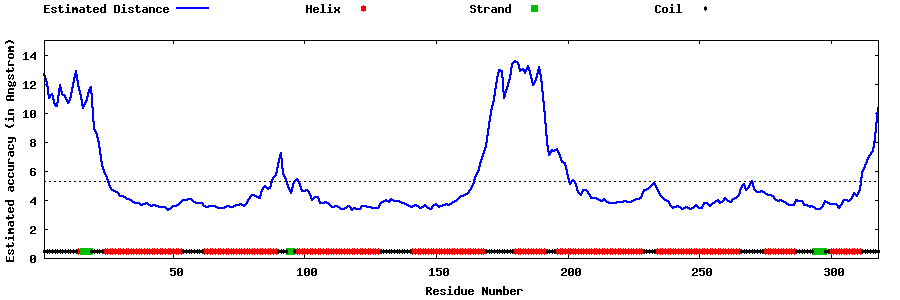 | |||
|
| |||
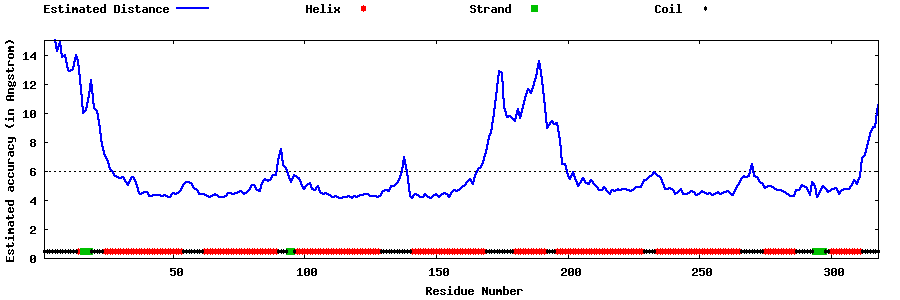 | |||
|
| |||
 | |||
|
| |||