

| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MLSAGLGLLMLVAVVEFLIGLIGNGSLVVWSFREWIRKFNWSSYNLIILGLAGCRFLLQWLIILDLSLFPLFQSSRWLRYLSIFWVLVSQASLWFATFLSVFYCKKITTFDRPAYLWLKQRAYNLSLWCLLGYFIINLLLTVQIGLTFYHPPQGNSSIRYPFESWQYLYAFQLNSGSYLPLVVFLVSSGMLIVSLYTHHKKMKVHSAGRRDVRAKAHITALKSLGCFLLLHLVYIMASPFSITSKTYPPDLTSVFIWETLMAAYPSLHSLILIMGIPRVKQTCQKILWKTVCARRCWGP | |
| CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCHHHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCCCCCCCC | |
| 98689999999999999999999899999989999817988767799999999999999999976872277277179999999999980999999999999987245389977999999872442999999999999999988774104678886204430331578999999999999999999999999999999999860879999999809999999999999999999999999999998715538999999999998488877999618709999999999867655678894 |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | |
| | | | | | | | | | | | | | | | |
| MLSAGLGLLMLVAVVEFLIGLIGNGSLVVWSFREWIRKFNWSSYNLIILGLAGCRFLLQWLIILDLSLFPLFQSSRWLRYLSIFWVLVSQASLWFATFLSVFYCKKITTFDRPAYLWLKQRAYNLSLWCLLGYFIINLLLTVQIGLTFYHPPQGNSSIRYPFESWQYLYAFQLNSGSYLPLVVFLVSSGMLIVSLYTHHKKMKVHSAGRRDVRAKAHITALKSLGCFLLLHLVYIMASPFSITSKTYPPDLTSVFIWETLMAAYPSLHSLILIMGIPRVKQTCQKILWKTVCARRCWGP | |
| 73432332112213312331331122002100110045340320110000003111100212333000000013320000000001221220000000000000000022311000001330431001203312231231111010313433331222323343210111133333333333323313320220022023202432443411304002200100220233233223113311112333442211111222102100000202404143013001300220202335578 |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSSSCCCCHHHHHHHHHHHCHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCSSSSCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCHHHHHHHHHHHHHHHHHHHHHHHCCCHHHHHHHHHHHHHCCCCCCCCCC MLSAGLGLLMLVAVVEFLIGLIGNGSLVVWSFREWIRKFNWSSYNLIILGLAGCRFLLQWLIILDLSLFPLFQSSRWLRYLSIFWVLVSQASLWFATFLSVFYCKKITTFDRPAYLWLKQRAYNLSLWCLLGYFIINLLLTVQIGLTFYHPPQGNSSIRYPFESWQYLYAFQLNSGSYLPLVVFLVSSGMLIVSLYTHHKKMKVHSAGRRDVRAKAHITALKSLGCFLLLHLVYIMASPFSITSKTYPPDLTSVFIWETLMAAYPSLHSLILIMGIPRVKQTCQKILWKTVCARRCWGP | |||||||||||||||||||||||||
| 1 | 4djhA | 0.13 | 0.20 | 0.90 | 1.38 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCPVKA-----LDFRTPLKAKIINICIWLLSSSVGISAIVGGTKVREDVDVIECSLSWWDLFMKICVFIFAVIPVLIIIVCYTLMILRLKSVRLLSDR-----------NLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----------P | |||||||||||||||||||
| 2 | 5tjvA | 0.09 | 0.21 | 0.91 | 2.54 | Download | LNPSQQLAIAVLSLTLGTFTVLENLLVLCVILH--SRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFDSRNVFLFKLGGVTASFTASVGSLFLAAIDRYISIHRPLAYKRIVTRPK----AVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGV-TSVLLLFIVYAYMYILWKAGKRAMSFSD--------QARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMF------------ | |||||||||||||||||||
| 3 | 4xt1A | 0.13 | 0.17 | 0.89 | 2.10 | Download | VLNQSKPVTLFLYGVVFLFGSIGNFLVIFTITW---RRRIQCSGDVYFINLAAADLLFVCTL---PLWMQYLLDSVPCTLLTACFYVAMFASLCFITEIALDRYYAIVYMRYRP--------VKQACLFSIFWWIFAVIIAIPHFMVVTKKDNQCMTDYLEVSYPIILNVELMLGAFVIPLSVISYCYYRISRIVA-------VSQSRH---KGRIVRVLIAVVLVFIIFWLPYHLTLFVDTLKLLISSSKRALILTESLAFCHCCLNPLLYVFVGTKFRQELHCLLAEFR-------- | |||||||||||||||||||
| 4 | 4djh | 0.12 | 0.15 | 0.93 | 1.55 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR---YTKMKTATNIYIFNLALADALVTTT-MPFQSTVYLNWPDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKA---L-DFRTPLKAKIINICIWLLSSSVGISAIVLGG-TKVREDVDVIECSLQDDFMICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSGNIFTDAYREKDRNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 5 | 5glh | 0.13 | 0.17 | 0.89 | 1.16 | Download | IKETFKYINTVVSCLVFVLGIIGNSTLLYIIYK--------NGPNILIASLALGDLLHIVIAIPINVYKLLPFGAEMCKLVPFIQKASVGITVLSLCALSIDRYRAVAS---W-----------WTAVEIVLIWVVSVVLAVPEAIGFDIMYKGSYLRICLQKAFMQFDWWLFSFYFCLPLAITAFFYTLMTCEMLRKNEGLRLTWDAYLNDHLKQRREVAKTVFCLVLVFALCWLPLHLARILKTLYNNLVLDYIGINMASLNSCANPIALYLVSKRFKNAFKSAL------------ | |||||||||||||||||||
| 6 | 4djhA | 0.12 | 0.20 | 0.91 | 1.49 | Download | ---AIPVIITAVYSVVFVVGLVGNSLVMFVIIRY---TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMNFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVKALDFRTPLKAKIINICIWLLSSSVGISAIVLGGTKVREDVDVIECSLQYSWWDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGAALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCFP---------- | |||||||||||||||||||
| 7 | 3uon | 0.13 | 0.14 | 0.93 | 1.73 | Download | ----EVVFIVLVAGSLSLVTIIGNILVMVSIKV---NRHLQTVNNYFLFSLACADLIIGVFSMNLYTIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVVEDGECYIQFF--SNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINIFAAKGTWDAYPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM----------- | |||||||||||||||||||
| 8 | 4n6hA | 0.11 | 0.21 | 0.93 | 2.34 | Download | SSLALAIAITALYSAVCAVGLLGNVLVMFGIVRY---TKMKTATNIYIFNLALADALATSTLPFQSAKYLMEFGELLCKAVLSIDYYNMFTSIFTLTMMSVDRYIAVCHPVK----ALDFRTPAKAKLINICIWVLASGVGVPIMVMAVTRPRDGAVVCMLQYWDTVTKICVFLFAFVVPILIITVCYGLMLLRLRSV------RLLSGSKEKDRSLRRITRMVLVVVGAFVVCWAPIHIFVIVWTLVDIVAALHLCIALGYANSSLNPVLYAFLDENFKRCFRQLCRK--PCG----- | |||||||||||||||||||
| 9 | 4djhA | 0.13 | 0.20 | 0.90 | 1.41 | Download | -SPAIPVIITAVYSVVFVVGLVGNSLVMFVIIR----TKMKTATNIYIFNLALADALVTTTMPFQSTVYLMPFGDVLCKIVLSIDYYNMFTSIFTLTMMSVDRYIAVC---HPVKA-LDFRTPLKAKIINICIWLLSSSVGISAIVLKVREDVDVIECSLQFPDDLFMKICVFIFAFVIPVLIIIVCYTLMILRLKSVRLLSD-----------RNLRRITRLVLVVVAVFVVCWTPIHIFILVEALGSALSSYYFCIALGYTNSSLNPILYAFLDENFKRCFRDFCF----------P | |||||||||||||||||||
| 10 | 2z73A | 0.12 | 0.20 | 0.97 | 1.56 | Download | VPDAVYYSLGIFIGICGIIGCGGNGIVIYLFTKT---KSLQTPANMFIINLAFSDFTFSLVNGFPLMTISCFLKKAACKVYGFIGGIFGFMSIMTMAMISIDRYNVIGPMAASKKMS-----HRRAFIMIIFVWLWSVLWAIGPIFGWGATLEGVLCNCSFDYITRSNILCMFILGFFGPILIIFFCYFNIVMSVSNHEKEMAAMAKRLNAKEANAEMRLAKISIVIVSQFLLSWSPYAVVALLAQFGPTPYAAQLPVMFAKASAIHNPMIYSVSHPKFREAISQTFPWVLTCCQFDDK | |||||||||||||||||||
| ||||||||||||||||||||||||||
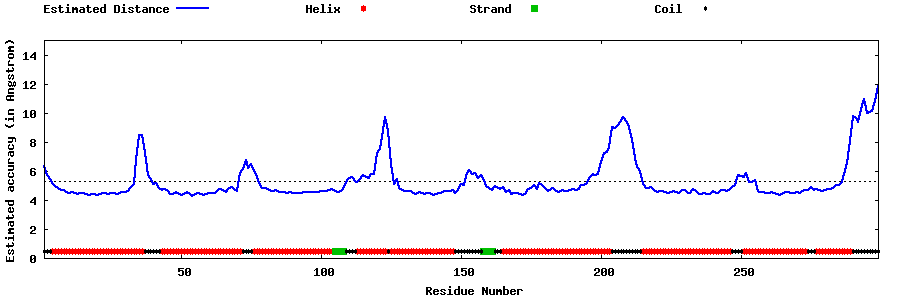
| Generated 3D models | Estimated local accuracy of models | ||
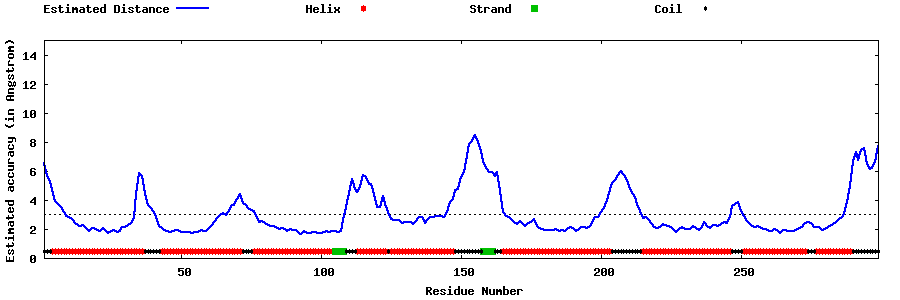 | |||
|
|
|||
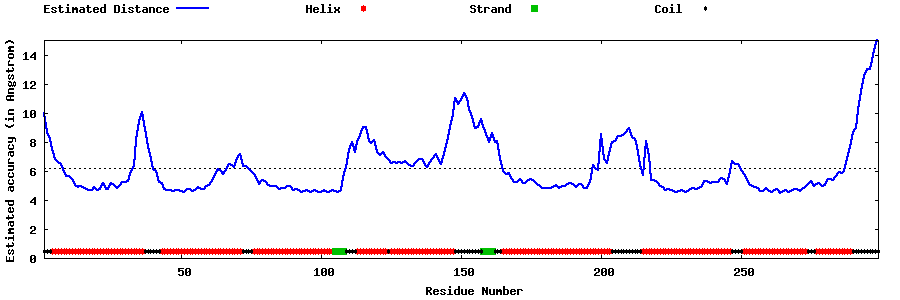 | |||
|
| |||
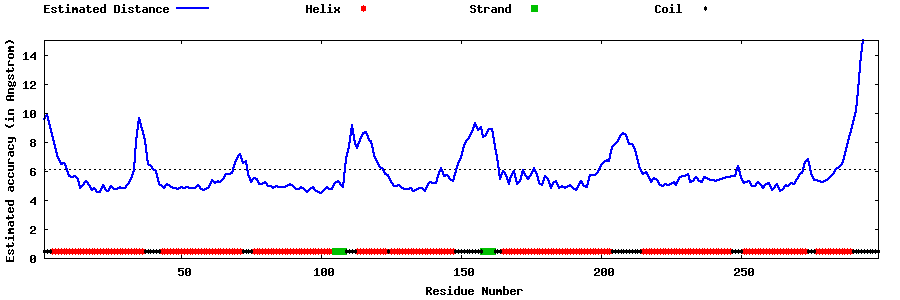 | |||
|
| |||
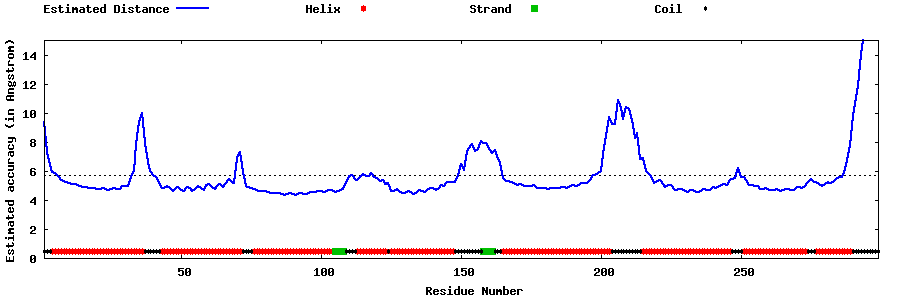 | |||
|
| |||
 | |||
|
| |||