GPCR-I-TASSER results for Q8NH92
[Click on Q8NH92_results.tar.bz2 to download the tarball file including all modeling results listed on this page]
| Submitted Sequence |
| >Q8NH92 MKTFSSFLQIGRNMHQGNQTTITEFILLGFFKQDEHQNLLFVLFLGMYLVTVIGNGLIIV AISLDTYLHTPMYLFLANLSFADISSISNSVPKMLVNIQTKSQSISYESCITQMYFSIVF VVIDNLLLGTMAYDHFVAICHPLNYTILMRPRFGILLTVISWFLSNIIALTHTLLLIQLL FCNHNTLPHFFCDLAPLLKLSCSDTLINELVLFIVGLSVIIFPFTLSFFSYVCIIRAVLR VSSTQGKWKAFSTCGSHLTVVLLFYGTIVGVYFFPSSTHPEDTDKIGAVLFTVVTPMINP FIYSLRNKDMKGALRKLINRKISSL |
| Predicted Secondary Structure |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | MKTFSSFLQIGRNMHQGNQTTITEFILLGFFKQDEHQNLLFVLFLGMYLVTVIGNGLIIVAISLDTYLHTPMYLFLANLSFADISSISNSVPKMLVNIQTKSQSISYESCITQMYFSIVFVVIDNLLLGTMAYDHFVAICHPLNYTILMRPRFGILLTVISWFLSNIIALTHTLLLIQLLFCNHNTLPHFFCDLAPLLKLSCSDTLINELVLFIVGLSVIIFPFTLSFFSYVCIIRAVLRVSSTQGKWKAFSTCGSHLTVVLLFYGTIVGVYFFPSSTHPEDTDKIGAVLFTVVTPMINPFIYSLRNKDMKGALRKLINRKISSL | |
| CCCHHHHHHHHHHCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSCCHCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSCCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCCCC | |
| 9413565677522647688606535211699882156899999999999999989999999962886556299998889998653341588998899844897475898999999999999989999999987515501665437854376089999999999999999999999863058999935893678699875616442688999999999999987999999999999884113678986751057077899999985713246706898788988828998401646254446423660889999999984110779 |
| Predicted Solvent Accessibility |
| 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | MKTFSSFLQIGRNMHQGNQTTITEFILLGFFKQDEHQNLLFVLFLGMYLVTVIGNGLIIVAISLDTYLHTPMYLFLANLSFADISSISNSVPKMLVNIQTKSQSISYESCITQMYFSIVFVVIDNLLLGTMAYDHFVAICHPLNYTILMRPRFGILLTVISWFLSNIIALTHTLLLIQLLFCNHNTLPHFFCDLAPLLKLSCSDTLINELVLFIVGLSVIIFPFTLSFFSYVCIIRAVLRVSSTQGKWKAFSTCGSHLTVVLLFYGTIVGVYFFPSSTHPEDTDKIGAVLFTVVTPMINPFIYSLRNKDMKGALRKLINRKISSL | |
| 6430221131254047503030010000000432511210023023112210332320020000003000000000100001031110033020000011443302030020011000000110000000000000000021020001001200000012012100100310010000010047240100001132004000120210001012212331331230033022100000010314612310000000000000011100100001022233443320000000333033102000010320130022004333216 | |
| Values range from 0 (buried residue) to 9 (highly exposed residue) | |
| Predicted normalized B-factor |
| Top 10 templates used by GPCR-I-TASSER |
| Rank | PDB Hit | Iden1 | Iden2 | Cov. | Norm. Z-score | Download Align. | 20 40 60 80 100 120 140 160 180 200 220 240 260 280 300 320 | | | | | | | | | | | | | | | | | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sec.Str Seq | CCCHHHHHHHHHHCCCCCCCCCHHHHSSCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCHHHHHHHHHHHHHHHHCCCHHHHHHHHHCCCCSSCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCCCSSCCHCHHHHHHHHHHHHHHHHHHHHHHHHHHCCCCCCCSSCCCCCCHHHHHHHCCCCCHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHHSSCCCCCCCCCCCCCCHHHHHHHHHHHCCHHSSSCCCCCCCCCCCCSSSSSSCCCCCCCCCCHHHCCCCHHHHHHHHHHHHHHCCCC MKTFSSFLQIGRNMHQGNQTTITEFILLGFFKQDEHQNLLFVLFLGMYLVTVIGNGLIIVAISLDTYLHTPMYLFLANLSFADISSISNSVPKMLVNIQTKSQSISYESCITQMYFSIVFVVIDNLLLGTMAYDHFVAICHPLNYTILMRPRFGILLTVISWFLSNIIALTHTLLLIQLLFCNHNTLPHFFCDLAPLLKLSCSDTLINELVLFIVGLSVIIFPFTLSFFSYVCIIRAVLRVSSTQGKWKAFSTCGSHLTVVLLFYGTIVGVYFFPSSTHPEDTDKIGAVLFTVVTPMINPFIYSLRNKDMKGALRKLINRKISSL | |||||||||||||||||||||||||
| 1 | 3v2yA | 0.17 | 0.21 | 0.82 | 1.99 | Download | -------------------------------DKENSIKLTSVVFILICCFIILENIFVLLTIWKTKKFHRPMYYFIGNLALSDLLAGVAYTANLLLSGATTYKLTPAQWFLREGSMFVALS-ASVFSLLAIAIERYITMLKN--------NFRLFLLISACWVISLILGG--------LPIMGWNCISA--------LSSCSTVLPLYHKHYILFCTTVFTLLLLSIVILYCRIYSLVRTRNSRSSENALLKTVIIVLSVFIACWAPLFILLLLDVGCKVKTCDILFRLVLAVLNSGTNPIIYTLTNKEMRRAFIRIMGRPL--- | |||||||||||||||||||
| 2 | 4iaqA | 0.20 | 0.23 | 0.82 | 2.20 | Download | -------------------------YIYQDSISLPWKVLLVMLLALITLATTLSNAFVIATVYRTRKLHTPANYLIASLAVTDLLVSILVMPISTMYTVTGRWTLGQVVCDFWLSSDITCCTASIWHLCVIALDRYWAITDAVEYSAKRTPKRAAVMIALVWVFSISISL-----------------PPFFCVVNI-------------LYTVYSTVGAFYFPTLLLIALYGRIYVEARSRIIQKYLLMERKATKTLGIILGAFIVCWLPFFIISLVMPIHLAIFDFFTWLGYLNSLINPIIYTMSNEDFKQAFHKLIRFK---- | |||||||||||||||||||
| 3 | 3emlA | 0.22 | 0.21 | 0.86 | 3.61 | Download | ---------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAII-VGLFALCWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 4 | 3uonA | 0.19 | 0.23 | 0.85 | 2.51 | Download | ---------------------------------TFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIV--GVRTVEDGECYIQFF------S---NAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRINISREKKVTRTILAILLAFIITWAPYNVMVLINTFCAIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLM------ | |||||||||||||||||||
| 5 | 4bvnA | 0.19 | 0.21 | 0.84 | 2.05 | Download | --------------------------------LSQQEAGMSLLMALVVLLIVAGNVLVIAAIGSTQRLQTLTNLFITSLACADLVVGLLVVPFGATLVVRGTWLWGSFLCELWTSLDVLCVTASVETLCVIAIDRYLAITSPFRYQSLMTRARAKVIICTVWAISALVSFLPIMM----HWWRDEDPQALKCYQD----PGCCDFVTNRAYAIASSIISFYIPLLIMIFVALRVYREAKE-----REHKALKTLGIIMGVFTLCWLPFFLVNIVNVFNRVPKWLFVAFNWLGYANSAMNPIILC-RSPDFRKAFKRLLA------ | |||||||||||||||||||
| 6 | 3emlA | 0.22 | 0.21 | 0.85 | 3.96 | Download | ----------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLGWNNCGGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHIINCF-TFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 7 | 2r4rA | 0.23 | 0.19 | 0.62 | 2.97 | Download | ----------------------------------------GIVMSLIVLAIVFGNVLVITAIA-FERLQTVTNYFITSLACADLVMGLAVVPFG-----------------FWTSIDVLCVTASIETLCVIAVDRYFAITSPFKYQSLLTKNKARVIILMVWIVSGLTS----------------------------------------------SIVSFYVPLVIMVFVYSRVFQEAKRIDKSLKEHKALKTLGIIMGTFTLCWLPFFI-----------------LNWIGYVNSGFNPLIYC-RSPDFRIAFQELLCLRRSS- | |||||||||||||||||||
| 8 | 1u19A | 0.15 | 0.20 | 0.92 | 2.56 | Download | -------------MNGSNKTGV-----VRSPAEPWQFSMLAAYMFLLIMLGFPINFLTLYVTVQHKKLRTPLNYILLNLAVADLFMVFGGFTTTLYTSLHGYFVFGPTGCNLEGFFATLGGEIALWSLVVLAIERYVVVCKPMSNFRFGENHAIMGVAFTWVMALACAAPPLVGWSRYIPEGMQCSCGIDYYT-------PHEETNNESFVIYMFVVHF-IIPLIVIFFCYGQLVFTVKEAAAQQQESRMVIIMVIAFLICWLPYAGVAFYIFTHQGSDFGPIFMTIPAFFAKTSAVYNPVIYIMMNKQFRNCMVTTLCCGKNPL | |||||||||||||||||||
| 9 | 3emlA | 0.22 | 0.21 | 0.86 | 5.55 | Download | ---------------------------------IMGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAITIST--GFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVP-----------MNYMVYFNFFACVLVPLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFALCWLPLHI-INCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| 10 | 3emlA | 0.23 | 0.21 | 0.86 | 5.45 | Download | I---------------------------------MGSSVYITVELAIAVLAILGNVLVCWAVWLNSNLQNVTNYFVVSLAAADIAVGVLAIPFAIT--ISTGFCAACHGCLFIACFVLVLTQSSIFSLLAIAIDRYIAIRIPLRYNGLVTGTRAKGIIAICWVLSFAIGLTPMLNCGQSQGCGEGQVACLFEDVVPMNYMVYFNFFACVLV-----------PLLLMLGVYLRIFLAARRQLVHAAKSLAIIVGLFAL-CWLPLHIINCFTFFCPDCSHAPLWLMYLAIVLSHTNSVVNPFIYAYRIREFRQTFRKIIRSHVLRQ | |||||||||||||||||||
| ||||||||||||||||||||||||||
| Top 5 Models predicted by GPCR-I-TASSER |
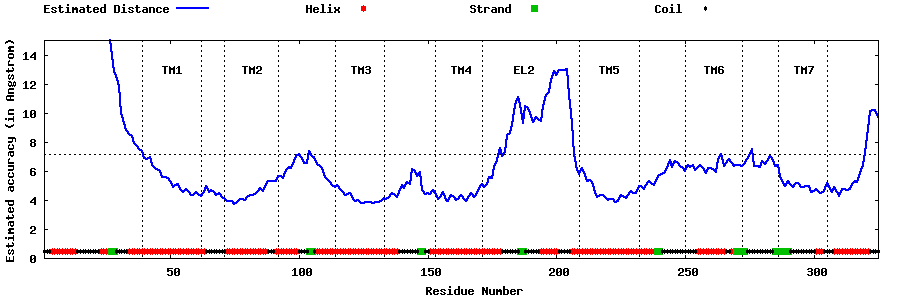
| Generated 3D models | Estimated local accuracy of models | ||
 | |||
|
|
|||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
 | |||
|
| |||
| Please cite following articles when you use the GPCR-I-TASSER server: | |
| J Zhang, J Yang, R Jang, Y Zhang. Hybrid structure modeling of G protein-coupled receptors in the human genome, submitted (2015). |
