

 [Home]
[Forum]
[Example]
[Help]
[Home]
[Forum]
[Example]
[Help]
About I-TASSER-MR
I-TASSER-MR is developed to solve structures by molecular replacement by using iterative fragmental structure assembly simulations followed by progressive sequence truncation that trims off regions with high structural variation.
User Input
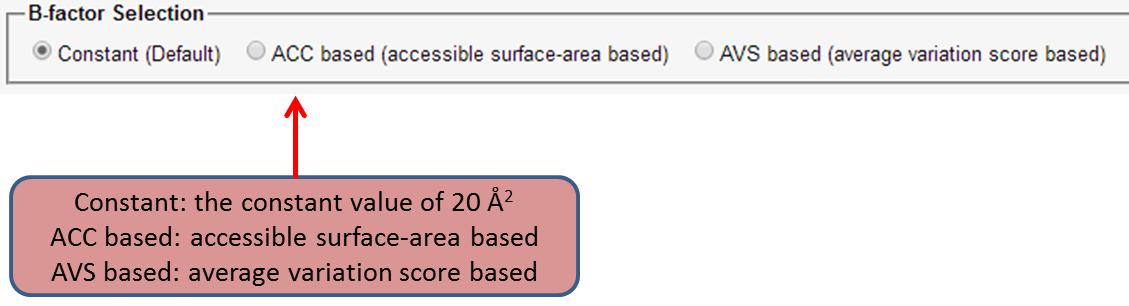
The user need to upload the amino acid sequence of the query protein, copy number(i.e. the number of monomers in asymmetric unit), and the crystallographic structure factors in MTZ format. The users are allowed to choose to use the three different B factors.
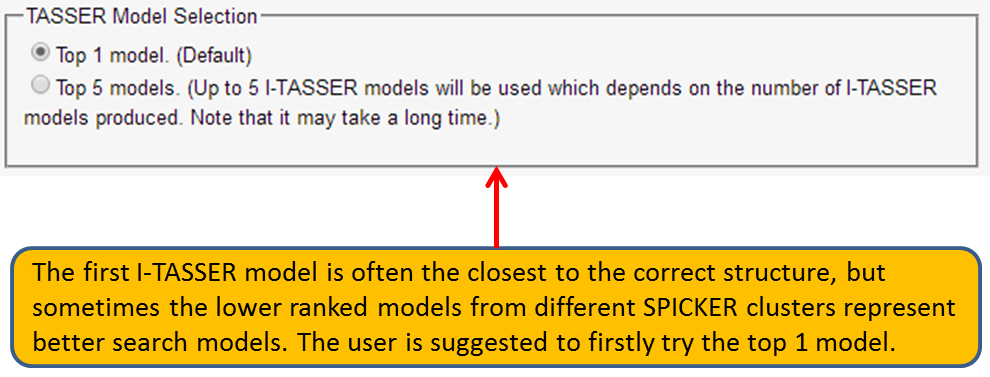
The users are also allowed to use the first or all the top 5 I-TASSER models for further MR process and refinement.
Providing an e-mail address is recommended for recieving the prediction results, else the user may opt to bookmark the web-link provided at the end of submission.
Output
The server outputs include:
(i)the summary of the query,
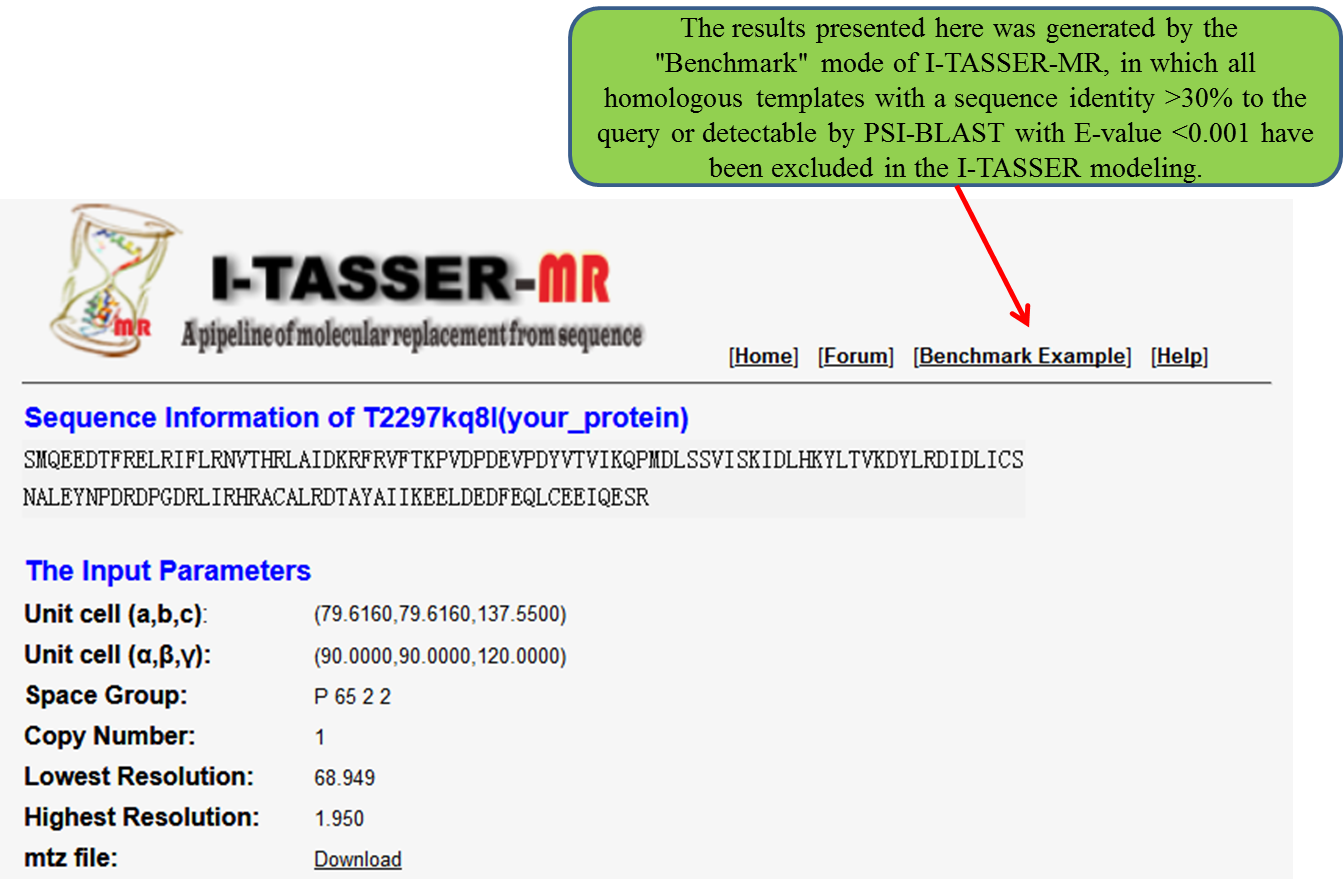
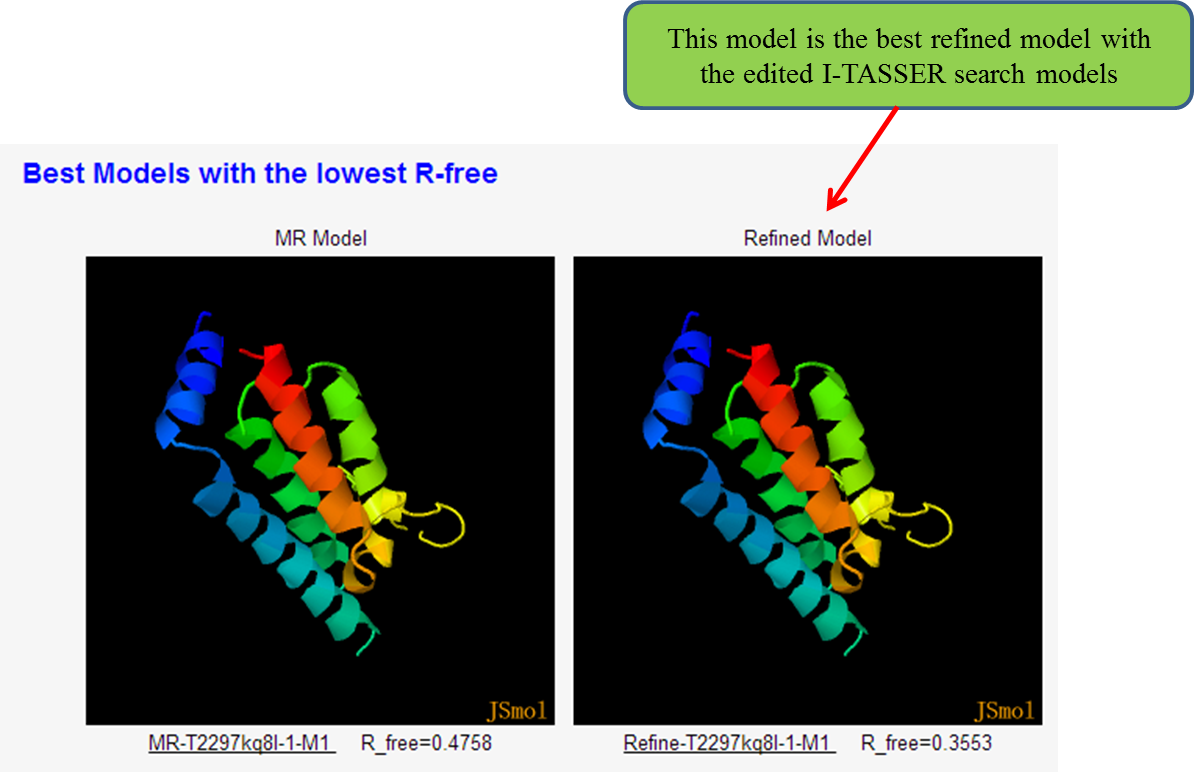
(ii) the best refined model and the corresponding MR model
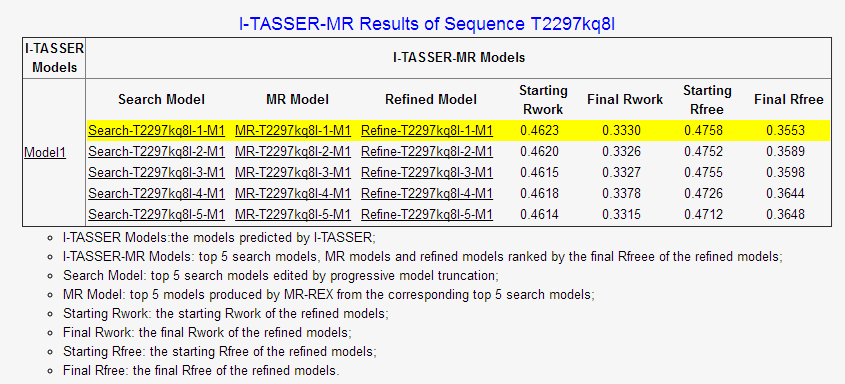
(iii) the top 5 search models, MR models and refined models ranked by Rfree of the refined models,
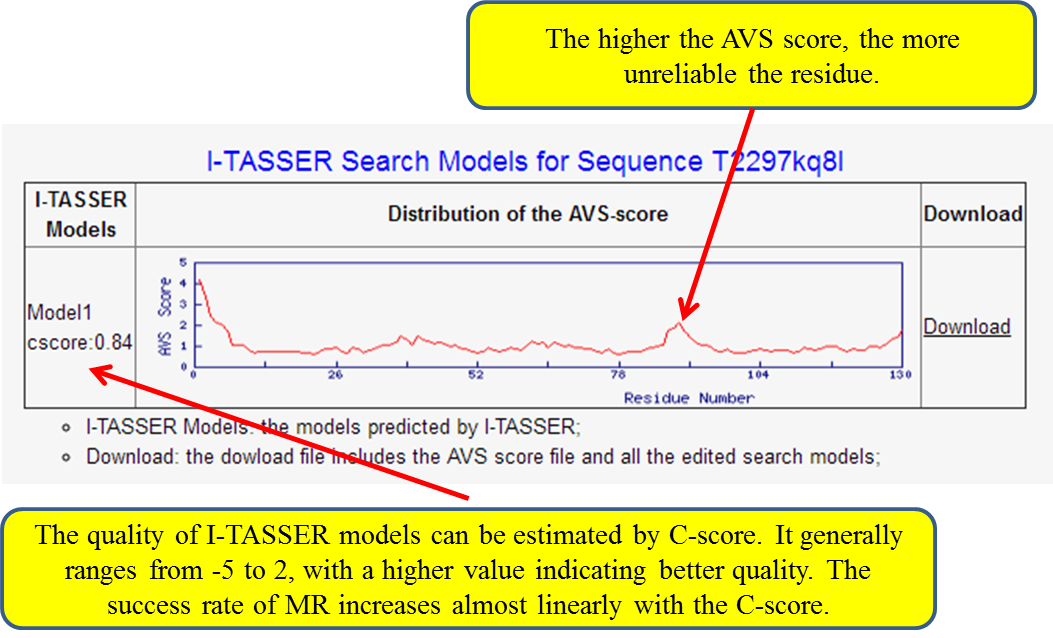
(iv) Search models produced according to the AVS-score,
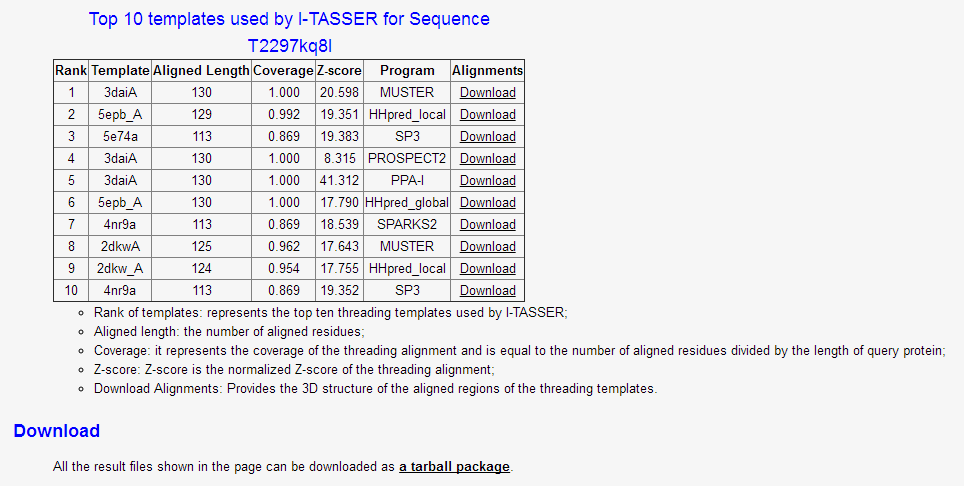
(v) the top 10 templates used by I-TASEER for the query protein sequence
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218