

| What is the SAXSTER server? |
SAXSTER server is an internet service to couple the raw SAXS data with protein fold-recognition algorithms to improve template-based protein structure predictions.
| How does SAXSTER identify the best templates? |
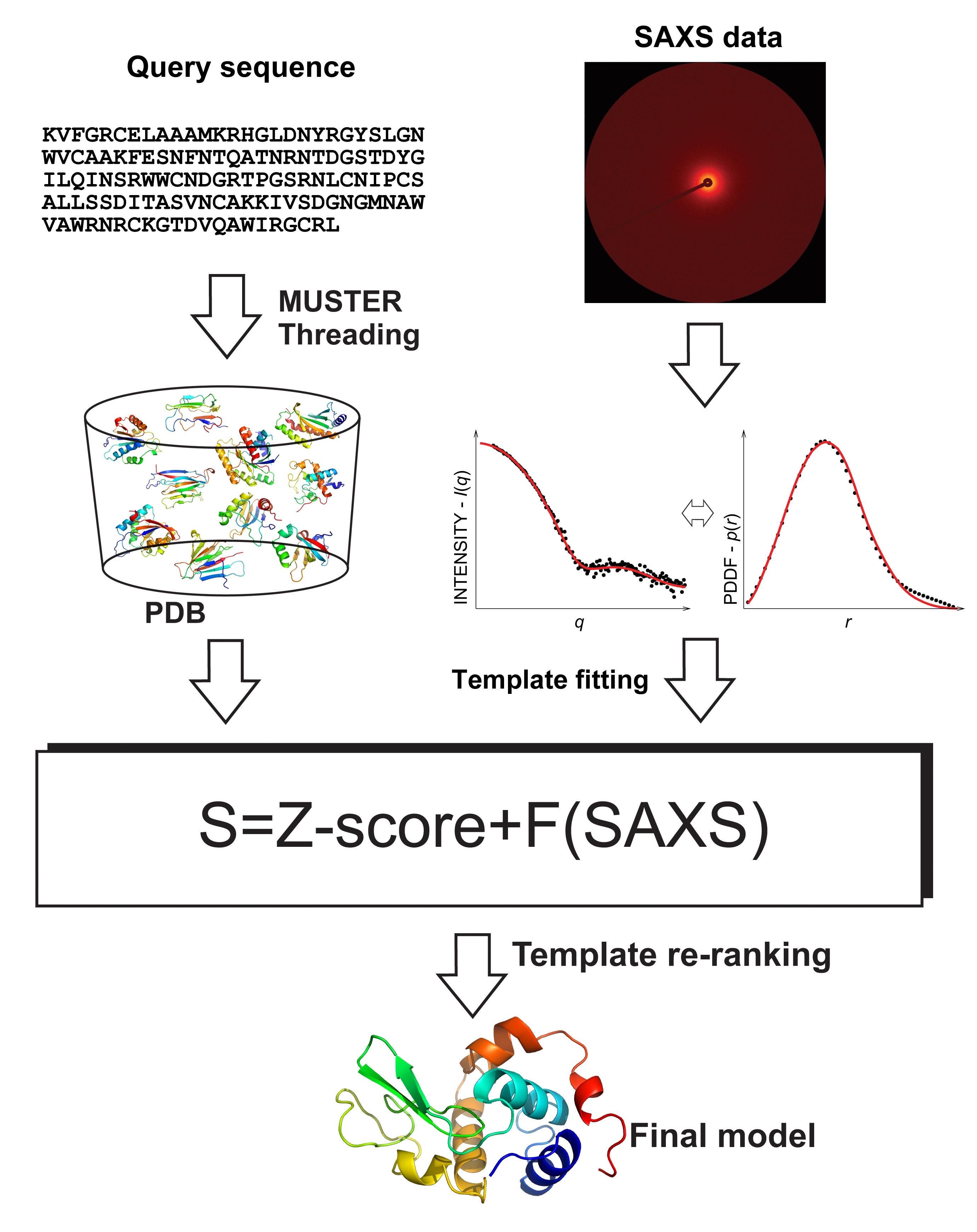
The SAXS-assisted MUSTER fold-recognition algorithm, SAXSTER, consists of five steps: threading-based template identification, full-length model construction by I-TASSER, coarse-grained SAXS profile calculation, SAXS data matching, and template re-ranking. The inputs of SAXSTER include the amino acid sequence of the target protein and its respective SAXS data (either intensity profile in reciprocal space or Pair Distance Distribution Function, PDDF, in real space), with the outputs being the Top 10 template structures and the query-template alignments (see Figure 1). The SAXSTER server also provides full-atomic models built by MODELLER from the Top 10 alpha-Carbon trace generated by I-TASSER from the threading alignments.
| How to cite SAXSTER |
You are requested to cite following article when you use the SAXSTER server:
| Contact information |
The SAXSTER server is in active development with the goal to provide the reliable structure predictions. Please help us achieve the goal by sending your questions, feedback, and comments to: yangzhanglab@umich.edu.
yangzhanglab![]() umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218
umich.edu
| (734) 647-1549 | 100 Washtenaw Avenue, Ann Arbor, MI 48109-2218