| Synonyms | Ratacand
BCP01137
SMR002203608
C-266
Candesartan [BAN]
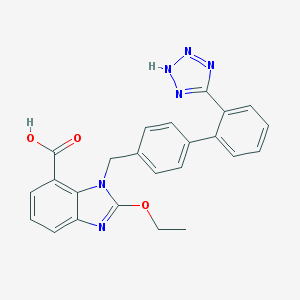
2-Ethoxy-1-((2-(1H-tetrazole-5-yl)biphenyl-4-yl)methyl)-1H-benzamidazole-7-carboxylic acid.
2-ethoxy-1-[[2'-(1h-tetrazol-5-yl)(1,1'-biphenyl)-4-yl]methyl]-1h-benzimidazole-7-carboxylic acid
D00522
2-ethoxy-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4ethyl]}-1H-benzimidazole-7-carboxylic acid
DTXSID0022725
2-ethoxy-3-[[4-[2-(1H-tetrazol-5-yl)phenyl]phenyl]methyl]benzimidazole-4-carboxylic acid
HMS3651C13
KB-48662
AB01275447_03
MolPort-005-943-739
AM84369
[3H]candesartan
ST2414902
API0001828
S-2839
BDBM50240609
SR-05000001447-1
1-((2'-(1H-tetrazol-5-yl)biphenyl-4-yl)methyl)-2-ethoxy-1H-benzo[d]imidazole-7-carboxylic acid
candesartan[candesartancilexetilintermediates]
2-ethoxy-1-({4-[2-(1H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl}methyl)-1H-1,3-benzodiazole-7-carboxylic acid
CS-2146
2-Ethoxy-1-[[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl]-benzimidazole-7-carboxylic acid
2-Ethoxy-3-[2''-(1H-tetrazol-5-yl)-biphenyl-4-ylmethyl]-3H-benzoimidazole-4-carboxylic acid(CV-11974)
FT-0083585
2-ethoxy-3-[[4-[2-(2H-tetrazol-5-yl)phenyl]phenyl]methyl]benzimidazole-4-carboxylic acid
HTQMVQVXFRQIKW-UHFFFAOYSA-N
A807545
L000156
AC1L1DWT
NCGC00167474-04
TL8000897
AX8018243
SC-15284
Blopress (TN)
Candesartan (USAN/INN)
1H-Benzimidazole-7-carboxylic acid, 2-ethoxy-1-((2'-(1H-tetrazol-5-yl)(1,1'-biphenyl)-4-yl)methyl)-
CCG-213059
2-ethoxy-1-<<2'-(1H-tetrazol-5-yl)biphenyl-4-yl>methyl>-1H-benzimidazole-7-carboxylic acid;
CV-11974
2-ethoxy-1-[[2'-(2h-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl]-1h-benzimidazole-7-carboxylic acid
DSSTox_GSID_22725
2-Ethoxy-3-[[4-[2-(1H-tetrazol-5-yl)phenyl]phenyl] methyl]-3H-benzoimidazole-4-carboxylic acid
GTPL6907
481C597
J-007281
AB01275447-01
MLS004774041
AKOS015888154
NSC759858
SR-05000001447-2
UNII-S8Q36MD2XX
Ratacand (TN)
BCP9000479
SMR003500711
C07468
1-((2'-(1H-Tetrazol-5-yl)-[1,1'-biphenyl]-4-yl)methyl)-2-ethoxy-1H-benzo[d]imidazole-7-carboxylic ac
Candesartan [USAN:INN:BAN]
2-Ethoxy-1-(p-(o-1H-tetrazol-5-ylphenyl)benzyl)-7-benzimidazolecarboxylic acid
CHEMBL1016
2-ethoxy-1-[[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl]-1H-benzimidazole-7-carboxylic acid
D0A0QL
2-ethoxy-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4ethyl}-1H-benzimidazole-7-carboxylic acid
EBD40593
2-ethoxy-3-[[4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl]methyl]benzimidazole-4-carboxylic acid
HMS3715F13
KS-000001UX
AB07470
NCGC00167474-01
Amias (TN)
SW199612-2
AT-3243
s1578
BIDD:GT0350
candesartan ( for candesartan cilexetil )
1-((2'-(2H-tetrazol-5-yl)-[1,1'-biphenyl]-4-yl)methyl)-2-ethoxy-1H-benzo[d]imidazole-7-carboxylic acid
CAS-139481-59-7
2-ethoxy-1-({4-[2-(2H-1,2,3,4-tetrazol-5-yl)phenyl]phenyl}methyl)-1H-1,3-benzodiazole-7-carboxylic acid
CTK8E8165
2-Ethoxy-1-[[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl]benzimidazole-7-carboxylic Acid
DB13919
2-Ethoxy-3-[2''-(2H-tetrazol-5-yl)-biphenyl-4-ylmethyl]-3H-benzoimidazole-4-carboxylic acid
FT-0602912
2-Ethoxy-3-[[4-[2-(2H-tetrazol-5-yl)phenyl]phenyl]methyl]benzimidazole-4-carboxylic acid.
HY-B0205
A808309
LS-32740
ACT02607
NCGC00167474-06
Tox21_112478
Pharmakon1600-01502288
BC205774
SCHEMBL3938
BR-57139
Candesartan - Atacand
2-(ethyloxy)-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl}-1H-benzimidazole-7-carboxylic acid
CHEBI:3347
2-Ethoxy-1-[[2'-(1 H-tetrazol-5-yl)biphenyl-4-yl]methyl]-benzimidazole-7-carboxylic acid
CV11974
2-ethoxy-1-{[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl}-1H-benzimidazole-7-carboxylic acid
DSSTox_RID_75453
2-ethoxy-3-[[4-[2-(1h-tetrazol-5-yl)phenyl]phenyl]methyl]-3h-benzoimidazole-4-carboxylic acid
HMS2089M22
4CH-015556
J10263
AB01275447_02
MolPort-003-845-570
AKOS025117340
ZINC3782818
SR-05000001447-5
RTR-005177
BCPP000302
SR-05000001447
C24H20N6O3
1-((2'-(1H-Tetrazol-5-yl)-[1,1'-biphenyl]-4-yl)methyl)-2-ethoxy-1H-benzo[d]imidazole-7-carboxylic acid
Candesartan [USAN:INN]
2-ethoxy-1-({2'-(1H-tetrazol-5-yl)[1,1'-biphenyl]-4-yl}methyl)-1H-benzimidazole-7-carboxylic acid
cilexitil
2-Ethoxy-1-[[2'-(1H-tetrazol-5-yl)biphenyl-4-yl]methyl]-1H-benzimidazole-7-carboxylic Acid (Candesartan)
D0D5SQ
2-ethoxy-3-[2''-(1H-tetrazol-5-yl)-biphenyl-4-ylmethyl]-3H-benzoimidazole-4-carboxylic acid
EC 604-138-8
2-ethoxy-3-[[4-[2-(2H-tetrazol-5-yl)phenyl]phenyl]methyl]-4-benzimidazolecarboxylic acid
HSDB 7520
KS-5003
AC-203
NCGC00167474-02
AN-839
TCV-116 (prodrug)
Atacand, Blopress, Amias, Ratacand,Candesartan
S8Q36MD2XX
Blopress
candesartan (Atacand)
139481-59-7
CC0016
2-ethoxy-1-<<2'-(1H-tetrazol-5-yl)biphenyl-4-yl>methyl>-1H-benzimidazole-7-carboxylic acid
CV 11974
2-ethoxy-1-[[2'-(1h-tetrazol-5-yl)[1,1'-biphenyl]-4-yl]methyl]-1h-benzimidazole-7-carboxylic acid
DSSTox_CID_2725
2-Ethoxy-3-[2'-(1H-tetrazol-5-yl)-biphenyl-4-ylmethyl]-3H-benzimidazole-4-carboxylic acid
GTPL587
2-ethoxy-7-carboxy-1-(2'-(1H-tetrazol-5-yl)biphenyl-4-yl)methylbenzimidazole
I01-0374
AB0011997
MLS003915631
AK-57139
NSC-759858
Tox21_112478_1 [ Show all ] |
|---|
| Inchi ID | InChI=1S/C24H20N6O3/c1-2-33-24-25-20-9-5-8-19(23(31)32)21(20)30(24)14-15-10-12-16(13-11-15)17-6-3-4-7-18(17)22-26-28-29-27-22/h3-13H,2,14H2,1H3,(H,31,32)(H,26,27,28,29) |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417