

You can:
| Name | Cannabinoid receptor 2 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CNR2 |
| Synonym | Peripheral cannabinoid receptor rCB2 hCB2 cannabinoid receptor 2 (macrophage) cannabinoid receptor 2 (spleen) [ Show all ] |
| Disease | Immune disorder Inflammatory bowel disease Inflammatory disease Neuropathic pain Osteoporosis [ Show all ] |
| Length | 360 |
| Amino acid sequence | MEECWVTEIANGSKDGLDSNPMKDYMILSGPQKTAVAVLCTLLGLLSALENVAVLYLILSSHQLRRKPSYLFIGSLAGADFLASVVFACSFVNFHVFHGVDSKAVFLLKIGSVTMTFTASVGSLLLTAIDRYLCLRYPPSYKALLTRGRALVTLGIMWVLSALVSYLPLMGWTCCPRPCSELFPLIPNDYLLSWLLFIAFLFSGIIYTYGHVLWKAHQHVASLSGHQDRQVPGMARMRLDVRLAKTLGLVLAVLLICWFPVLALMAHSLATTLSDQVKKAFAFCSMLCLINSMVNPVIYALRSGEIRSSAHHCLAHWKKCVRGLGSEAKEEAPRSSVTETEADGKITPWPDSRDLDLSDC |
| UniProt | P34972 |
| Protein Data Bank | 5zty |
| GPCR-HGmod model | P34972 |
| 3D structure model | This structure is from PDB ID 5zty. |
| BioLiP | BL0438927 |
| Therapeutic Target Database | T37693 |
| ChEMBL | CHEMBL253 |
| IUPHAR | 57 |
| DrugBank | BE0000095 |
| Name | Anandamide |
|---|---|
| Molecular formula | C22H37NO2 |
| IUPAC name | (5Z,8Z,11Z,14Z)-N-(2-hydroxyethyl)icosa-5,8,11,14-tetraenamide |
| Molecular weight | 347.543 |
| Hydrogen bond acceptor | 2 |
| Hydrogen bond donor | 2 |
| XlogP | 5.4 |
| Synonyms | Anandamide (20:4, n-6) Arachidonoylethanolamide (AEA) BSPBio_001533 CS-0002859 HMS3649B09 [ Show all ] |
| Inchi Key | LGEQQWMQCRIYKG-DOFZRALJSA-N |
| Inchi ID | InChI=1S/C22H37NO2/c1-2-3-4-5-6-7-8-9-10-11-12-13-14-15-16-17-18-19-22(25)23-20-21-24/h6-7,9-10,12-13,15-16,24H,2-5,8,11,14,17-21H2,1H3,(H,23,25)/b7-6-,10-9-,13-12-,16-15- |
| PubChem CID | 5281969 |
| ChEMBL | CHEMBL15848 |
| IUPHAR | 2364 |
| BindingDB | 22988 |
| DrugBank | N/A |
Structure | 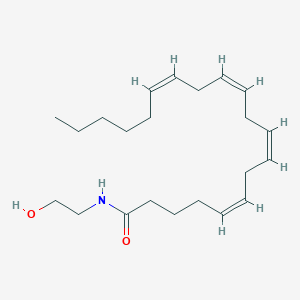 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Ki | <1000.0 nM | PMID11741470 | BindingDB,ChEMBL |
| Ki | 168.0 nM | PMID19361197 | BindingDB,ChEMBL |
| Ki | 180.0 nM | PMID19161308, PMID16682198, PMID16570929, PMID16213718 | BindingDB,ChEMBL |
| Ki | 306.0 nM | PMID10688601 | BindingDB |
| Ki | 371.0 nM | PMID23865723 | BindingDB,ChEMBL |
| Ki | 398.107 - 1995.26 nM | PMID7565624, PMID8819477, PMID7605349 | IUPHAR |
| Log Ki | 2.57 nM | PMID10882356 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417