

You can:
| Name | Cannabinoid receptor 1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CNR1 |
| Synonym | CB1 Central cannabinoid receptor SKR6R THC receptor CB1R [ Show all ] |
| Disease | Obesity; Diabetes Chemotherapy-induced nausea Diabetes; Obesity Drug abuse Hypertension; Diabetes; Obesity [ Show all ] |
| Length | 472 |
| Amino acid sequence | MKSILDGLADTTFRTITTDLLYVGSNDIQYEDIKGDMASKLGYFPQKFPLTSFRGSPFQEKMTAGDNPQLVPADQVNITEFYNKSLSSFKENEENIQCGENFMDIECFMVLNPSQQLAIAVLSLTLGTFTVLENLLVLCVILHSRSLRCRPSYHFIGSLAVADLLGSVIFVYSFIDFHVFHRKDSRNVFLFKLGGVTASFTASVGSLFLTAIDRYISIHRPLAYKRIVTRPKAVVAFCLMWTIAIVIAVLPLLGWNCEKLQSVCSDIFPHIDETYLMFWIGVTSVLLLFIVYAYMYILWKAHSHAVRMIQRGTQKSIIIHTSEDGKVQVTRPDQARMDIRLAKTLVLILVVLIICWGPLLAIMVYDVFGKMNKLIKTVFAFCSMLCLLNSTVNPIIYALRSKDLRHAFRSMFPSCEGTAQPLDNSMGDSDCLHKHANNAASVHRAAESCIKSTVKIAKVTMSVSTDTSAEAL |
| UniProt | P21554 |
| Protein Data Bank | 5tjv, 5u09, 5xr8, 5xra, 6n4b, 5tgz |
| GPCR-HGmod model | P21554 |
| 3D structure model | This structure is from PDB ID 5tjv. |
| BioLiP | BL0384680, BL0364157, BL0384679, BL0384681, BL0384682, BL0384683, BL0384684, BL0440253, BL0440254,BL0440255, BL0363267, BL0361447, BL0361446 |
| Therapeutic Target Database | T76685 |
| ChEMBL | CHEMBL218 |
| IUPHAR | 56 |
| DrugBank | BE0000061 |
| Name | CP55940 |
|---|---|
| Molecular formula | C24H40O3 |
| IUPAC name | 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol |
| Molecular weight | 376.581 |
| Hydrogen bond acceptor | 3 |
| Hydrogen bond donor | 3 |
| XlogP | 6.1 |
| Synonyms | 86687-39-0 CCG-204284 CP 868388 D0D6OH KFY70972J5 [ Show all ] |
| Inchi Key | YNZFFALZMRAPHQ-SYYKKAFVSA-N |
| Inchi ID | InChI=1S/C24H40O3/c1-4-5-6-7-14-24(2,3)19-11-13-21(23(27)16-19)22-17-20(26)12-10-18(22)9-8-15-25/h11,13,16,18,20,22,25-27H,4-10,12,14-15,17H2,1-3H3/t18-,20-,22-/m1/s1 |
| PubChem CID | 104895 |
| ChEMBL | CHEMBL559612 |
| IUPHAR | 730, 734 |
| BindingDB | 50072775 |
| DrugBank | N/A |
Structure | 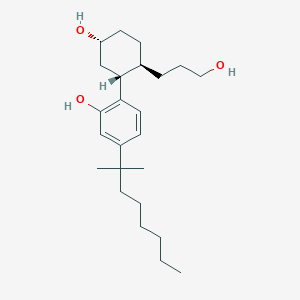 |
| Lipinski's druglikeness | This ligand has a partition coefficient log P greater than 5. |
| Parameter | Value | Reference | Database source |
|---|---|---|---|
| Activity | 100.0 % | PMID19500981 | ChEMBL |
| Activity | 104.3 % | PMID26035635, PMID21667972 | ChEMBL |
| Activity | 161.0 % | MedChemComm, (2014) 5:5:632 | ChEMBL |
| Activity | 228.0 % | PMID19278853 | ChEMBL |
| Bmax | 1.0 pmol/mg | PMID17004712 | ChEMBL |
| EC50 | 0.26 nM | PMID18680277 | BindingDB,ChEMBL |
| EC50 | 0.28 nM | PMID17630726 | BindingDB,ChEMBL |
| EC50 | 0.437 nM | PMID26203658 | BindingDB |
| EC50 | 0.437 nM | PMID26203658 | ChEMBL |
| EC50 | 0.631 nM | PMID21885167 | ChEMBL |
| EC50 | 0.7 nM | PMID23085772 | BindingDB,ChEMBL |
| EC50 | 1.259 nM | PMID20688519 | ChEMBL |
| EC50 | 1.4 nM | PMID25065940 | BindingDB |
| EC50 | 1.43 nM | PMID25065940 | ChEMBL |
| EC50 | 2.28 nM | PMID18680277, PMID24900561, PMID22916707 | BindingDB,ChEMBL |
| EC50 | 2.3 nM | PMID24900561 | BindingDB |
| EC50 | 3.9 nM | PMID25072877 | BindingDB,ChEMBL |
| EC50 | 4.9 nM | PMID26529344 | ChEMBL |
| EC50 | 5.0 nM | PMID18666769 | BindingDB,ChEMBL |
| EC50 | 7.0 nM | PMID25644673 | ChEMBL |
| EC50 | 7.0 nM | PMID25644673 | BindingDB |
| EC50 | 7.9 nM | PMID26151231 | BindingDB |
| EC50 | 7.93 nM | PMID26151231 | ChEMBL |
| EC50 | 8.0 nM | PMID25096297 | ChEMBL |
| EC50 | 8.0 nM | PMID25096297 | BindingDB |
| EC50 | 10.3 nM | PMID19115816 | BindingDB,ChEMBL |
| EC50 | 12.88 nM | PMID25096297, PMID24984935 | ChEMBL |
| EC50 | 13.0 nM | PMID25096297, PMID24984935 | BindingDB |
| EC50 | 19.95 nM | PMID21316962, MedChemComm, (2010) 1:1:54, PMID21074434 | BindingDB,ChEMBL |
| EC50 | 25.12 nM | PMID22421020 | ChEMBL |
| EC50 | 158.49 nM | PMID17027269 | ChEMBL |
| Emax | -90.1 % | PMID26529344 | ChEMBL |
| Emax | 67.36 % | PMID25096297 | ChEMBL |
| Emax | 72.1 % | PMID25065940 | ChEMBL |
| Emax | 100.0 % | PMID18666769, PMID17630726, PMID24984935, PMID23085772, PMID19115816 | ChEMBL |
| Emax | 348.0 % | PMID25072877 | ChEMBL |
| IC50 | 0.13 nM | PMID20047779 | BindingDB,ChEMBL |
| IC50 | 0.21 nM | PMID23582449 | ChEMBL |
| IC50 | 0.31 nM | PMID23466604 | ChEMBL |
| IC50 | 0.77 nM | PMID17630726 | BindingDB,ChEMBL |
| IC50 | 0.9 nM | PMID26988801, PMID27876250 | ChEMBL |
| IC50 | 1.4 nM | PMID18983139 | ChEMBL |
| IC50 | 2.1 nM | PMID17884496, PMID18006322 | BindingDB,ChEMBL |
| IC50 | 2.5 nM | PMID17027269 | BindingDB |
| IC50 | 2.512 nM | PMID17027269 | ChEMBL |
| IC50 | 9.0 nM | PMID26988801, PMID27876250 | BindingDB |
| IC50 | 14.0 nM | PMID10465552 | BindingDB |
| IC50 | 14.4 nM | PMID10465552 | ChEMBL |
| IC50 | 27.5 nM | PMID10465552 | ChEMBL |
| IC50 | 28.0 nM | PMID10465552 | BindingDB |
| Inhibition | 88.8 % | PMID23659286 | ChEMBL |
| Kd | 0.398107 - 3.16228 nM | PMID7775459, PMID8819477, PMID8526880, PMID1718258, PMID1331766, PMID8636122 | IUPHAR |
| Kd | 1.8 nM | PMID24092756 | BindingDB |
| Ki | 0.27 nM | PMID23466604 | ChEMBL |
| Ki | 0.316 nM | PMID21316962 | BindingDB |
| Ki | 0.3162 nM | PMID21316962, MedChemComm, (2010) 1:1:54, PMID21074434 | ChEMBL |
| Ki | 0.49 nM | PMID21867920 | BindingDB |
| Ki | 0.52 nM | PMID25072877 | BindingDB |
| Ki | 0.52 nM | PMID25072877 | ChEMBL |
| Ki | 0.58 nM | PMID23865723, PMID20218623 | BindingDB,ChEMBL |
| Ki | 0.58 nM | PMID23865723 | BindingDB |
| Ki | 0.6 nM | PMID21183257 | BindingDB,ChEMBL |
| Ki | 0.630958 - 5.01187 nM | PMID7565624, PMID8819477, PMID10188977 | IUPHAR |
| Ki | 1.2 nM | PMID18983139 | ChEMBL |
| Ki | 1.259 nM | PMID21885167 | ChEMBL |
| Ki | 1.28 nM | PMID24900561, PMID22916707 | BindingDB,ChEMBL |
| Ki | 1.3 nM | PMID24900561 | BindingDB |
| Ki | 1.36 nM | PMID20688519 | BindingDB,ChEMBL |
| Ki | 1.37 nM | PMID19278853 | ChEMBL |
| Ki | 1.37 nM | PMID19278853 | BindingDB |
| Ki | 5.2 nM | PMID11960486, PMID10465552 | BindingDB,ChEMBL |
| Ki | 13.0 nM | PMID16213718 | BindingDB,ChEMBL |
| Log Ki | -0.19 nM | PMID10882356 | ChEMBL |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417