| Synonyms | SR-01000883983-1
Sucrose ACS grade
Sucrose, analytical standard, for enzymatic assay kit SCA20
Sucrose, dust
Sucrose, Low Endotoxin, PharmaGrade, USP/NF, Ph Eur, Manufactured under appropriate GMP controls for pharma or biopharmaceutical production.
Sucrose, reagent grade
Sugar spheres (NF)
UNII-C151H8M554
HSDB 500
alpha-D-fructofuranosyl alpha-L-gulopyranoside
MolPort-003-926-939
alpha-D-Glucopyranosylbeta-D-fructofuranoside
Polysucrose
beta-D-fructofuranose-(2-1)-alpha-D-glucopyranoside
bmse000804
1-alpha-D-glucopyranosyl-2-beta-D-fructofranoside
CAS-57-50-1
131932-12-2
CRB0000841
2-{[3,4-dihydroxy-2,5-bis(hydroxymethyl)oxolan-2-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol
D(+)-Sucrose, ACS reagent
50857-68-6
DB02772
76056-38-7
FICOLL
87430-66-8
saccharose
Sucraloxum
Sucrose [USAN:JAN]
Sucrose, BioReagent, suitable for cell culture, suitable for insect cell culture, >=99.5% (GC)
Sucrose, for microbiology, ACS reagent, >=99.0%
Sucrose, NIST(R) SRM(R) 17f, optical rotation
Sucrose, USP
Sugar,(S)
White sugar
a-D-Glucopyranosyl A-D-fructofuranoside
KB-303246
alpha-D-Glucopyranoside, beta-D-fructofuranosyl
NCGC00164248-03
b -D-Fructofuranosyl a-D-glucopyranoside
S0111
beta-D-Fructofuranosyl-alpha-D-glucopyranoside
(2R,3R,4S,5S,6R)-2-((2S,3S,4S,5R)-3,4-dihydroxy-2,5-bis(hydroxymethyl)tetrahydrofuran-2-yloxy)-6-(hydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol
C00089
104242-10-6
CHEBI:17992
1481578-88-4
D(+)-OaIC
29764-06-5
D-Saccharose
5903AE
DSSTox_GSID_21288
80165-03-3
Glc(alpha1->2beta)Fru
ZINC4217475
SC-81182
Sucrose (JP17/NF)
Sucrose, ACS reagent
Sucrose, British Pharmacopoeia (BP) Reference Standard
Sucrose, Grade II, plant cell culture tested
Sucrose, purified
Sucrose|?-D-Fructofuranosyl ?-D-glucopyranoside
Tox21_201397
Granulated sugar
AKOS024306988
MFCD00006626
alpha-D-Glucopyranosyl beta-D-fructofuranoside
NCI-C56597
bet a-D-fructofuranosyl-alpha-D-Glucopyranoside
BIS3391
(2R,3R,4S,5S,6R)-2-{[(2S,3S,4S,5R)-3,4-dihydroxy-2,5-bis(hydroxymethyl)oxolan-2-yl]oxy}-6-(hydroxymethyl)oxane-3,4,5-triol
Cane sugar
12040-73-2
Compressible sugar, United States Pharmacopeia (USP) Reference Standard
1af6
D(+)-Sucrose, 99+%, for biochemistry, DNAse, RNAse and protease free
47185-09-1
D08TCV
65545-99-5
EINECS 200-334-9
85456-51-5
ST50308554
Sucrose Biochemical grade
Sucrose, anhydrous, free-flowing, Redi-Dri(TM), ACS reagent
Sucrose, European Pharmacopoeia (EP) Reference Standard
Sucrose, LR
Sucrose, SAJ first grade
Sugar, compressible (NF)
UNII-YUH1C99228 component CZMRCDWAGMRECN-UGDNZRGBSA-N
880257-62-5
HY-B1779
alpha-D-Glc-(1-->2)-beta-D-Fru
NCGC00164248-01
Amerfand
Rock candy
beta-D-Fructofuranosyl alpha-D-glucopyranoside
(+)-Sucrose
bmse000918
1-alpha-D-glucopyranosyl-2-beta-D-fructofuranoside
CC-34575
146054-35-5
CS-0013810
220376-22-7
D-(+)-Saccharose
51909-69-4
degrees xIC
78654-77-0
Fructofuranoside, alpha-D-glucopyranosyl, beta-D
WURCS=2.0/2,2,1/[ha122h-2b_2-5][a2122h-1a_1-5]/1-2/a2-b1
Saccharum
Sucraloxum [INN-Latin]
Sucrose, >=99.5%
Sucrose, BioUltra, for molecular biology, >=99.5% (HPLC)
Sucrose, for molecular biology, >=99.5% (GC)
Sucrose, p.a., ACS reagent
Sucrose, Vetec(TM) reagent grade, 99%
Table sugar
AC1L1LK0
LS-229
alpha-D-Glucopyranoside, beta-D-fructofuranosyl, homopolymer
NCGC00254237-01
BDBM50108105
beta-D-Fruf-(2<->1)-alpha-D-Glcp
(2R,3R,4S,5S,6R)-2-[(2S,3S,4S,5R)-3,4-dihydroxy-2,5-bis(hydroxymethyl)oxolan-2-yl]oxy-6-(hydroxymethyl)oxane-3,4,5-triol
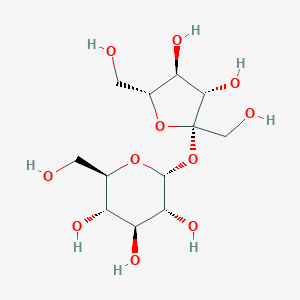
C12H22O11
1159795-78-4
CHEBI:65313
151756-02-4
D(+)-Saccharose
30027-72-6
D-Sucrose
635681-90-2
DSSTox_RID_76060
8027-47-2
GLC-(1-2)FRU
ZX-AFC000756
SR-01000883983
Sucrose (TN)
Sucrose, analytical standard
Sucrose, cell culture tested
Sucrose, JIS special grade
Sucrose, puriss., meets analytical specification of Ph. Eur., BP, NF
sugar
Tox21_300410
GTPL5411
alpha-(1R,2R,3S,4S,5R)-glucopyranosyl-beta-(2S,3S,4S,5R)-fructofuranoside
Microse
alpha-D-glucopyranosyl-(1->2)-beta-D-fructofuranoside
NSC 406942
beta-(2S,3S,4S,5R)-fructofuranosyl-alpha-(1R,2R,3S,4S,5R)-glucopyranoside
bmse000119
(alpha-D-Glucosido)-beta-D-fructofuranoside
Carbon isotopes in sucrose, NIST(R) RM 8542, IAEA-CH-6 sucrose
1206156-82-2
Confectioner's sugar
2-[(2S,3S,4S,5R)-2,5-bis(hydroxymethyl)-3,4-dihydroxyoxolan-2-yloxy](4S,5S,2R, 3R,6R)-6-(hydroxymethyl)-2H-3,4,5,6-tetrahydropyran-3,4,5-triol
D(+)-Sucrose, 99.7%, for biochemistry
47257-91-0
75398-84-4
Epitope ID:153236
86101-30-6
sacarosa
Sucralox
Sucrose [JAN:NF]
Sucrose, AR
Sucrose, for electrophoresis, >99%
Sucrose, meets USP testing specifications
Sucrose, United States Pharmacopeia (USP) Reference Standard
Sugar, confectioner's (NF)
White soft sugar (JP17)
92004-84-7
J-519846
alpha-D-Glc-(1-2)-beta-D-Fru
NCGC00164248-02
Amerfond
Rohrzucker
beta-D-fructofuranosyl-(2↔1)-alpha-D-glucopyranoside
(2R,3R,4S,5S,6R)-2-((2S,3S,4S,5R)-3,4-dihydroxy-2,5-bis(hydroxymethyl)tetrahydrofuran-2-ylhydroxymethyl)tetrahydro-2H-pyran-3,4,5-triol
C-30736
100405-08-1
CCRIS 2120
146187-04-4
CZMRCDWAGMRECN-UGDNZRGBSA-N
25702-74-3
D-(+)-Sucrose
57-50-1
DSSTox_CID_1288
786702-63-4
FT-0621912
Z1601554751
Sacharose
Sucrose, >=99.5% (GC)
Sucrose, BioXtra, >=99.5% (GC)
Sucrose, Grade I, plant cell culture tested
Sucrose, pure
Sucrose, Vetec(TM) reagent grade, RNase and DNase free
Tox21_112093
Glucopyranoside, beta-D-fructofuranosyl, alpha-D
AI3-09085
MCULE-5933491732
alpha-D-Glucopyranoside, beta-D-fructofuranosyl-
NCGC00258948-01
Beet sugar
BIA1125
(2R,3R,4S,5S,6R)-2-[(2S,3S,4S,5R)-3,4-dihydroxy-2,5-bis(hydroxymethyl)tetrahydrofuran-2-yl]oxy-6-(hydroxymethyl)tetrahydropyran-3,4,5-triol
C151H8M554
1192061-43-0
CHEMBL253582
1566560-95-9
D(+)-Sucrose
47167-52-2
D00025
64533-66-0
DTXSID2021288
8030-20-4 [ Show all ] |
|---|


![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417