

You can:
| Name | Muscarinic acetylcholine receptor M4 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM4 |
| Synonym | HM3 M4 receptor cholinergic receptor Chrm-4 cholinergic receptor, muscarinic 4 |
| Disease | Produce mydriasis and cycloplegia for diagnostic purposes Hypertension Irritable bowel syndrome Moderate and severe psychomotor agitation Mydriasis diagnosis [ Show all ] |
| Length | 479 |
| Amino acid sequence | MANFTPVNGSSGNQSVRLVTSSSHNRYETVEMVFIATVTGSLSLVTVVGNILVMLSIKVNRQLQTVNNYFLFSLACADLIIGAFSMNLYTVYIIKGYWPLGAVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPARRTTKMAGLMIAAAWVLSFVLWAPAILFWQFVVGKRTVPDNQCFIQFLSNPAVTFGTAIAAFYLPVVIMTVLYIHISLASRSRVHKHRPEGPKEKKAKTLAFLKSPLMKQSVKKPPPGEAAREELRNGKLEEAPPPALPPPPRPVADKDTSNESSSGSATQNTKERPATELSTTEATTPAMPAPPLQPRALNPASRWSKIQIVTKQTGNECVTAIEIVPATPAGMRPAANVARKFASIARNQVRKKRQMAARERKVTRTIFAILLAFILTWTPYNVMVLVNTFCQSCIPDTVWSIGYWLCYVNSTINPACYALCNATFKKTFRHLLLCQYRNIGTAR |
| UniProt | P08173 |
| Protein Data Bank | 5dsg |
| GPCR-HGmod model | P08173 |
| 3D structure model | This structure is from PDB ID 5dsg. |
| BioLiP | BL0339919,BL0339921, BL0339920 |
| Therapeutic Target Database | T20709, T50918 |
| ChEMBL | CHEMBL1821 |
| IUPHAR | 16 |
| DrugBank | BE0000405 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 441677 |  CHEMBL1602658 CHEMBL1602658 | C23H28N2O5S | 444.546 | 6 / 0 | 2.6 | Yes |
| 526 |  CHEMBL2407175 CHEMBL2407175 | C24H36N4O3 | 428.577 | 5 / 0 | 2.7 | Yes |
| 1009 |  CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1276 |  CHEMBL141736 CHEMBL141736 | C24H29N2O2+ | 377.508 | 2 / 0 | 2.8 | Yes |
| 1942 | 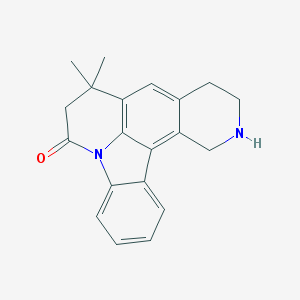 CHEMBL282874 CHEMBL282874 | C20H20N2O | 304.393 | 2 / 1 | 3.5 | Yes |
| 2272 | 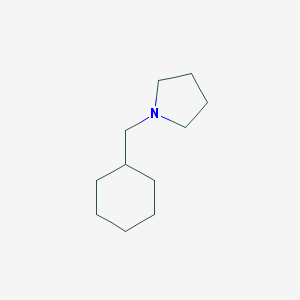 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2909 | 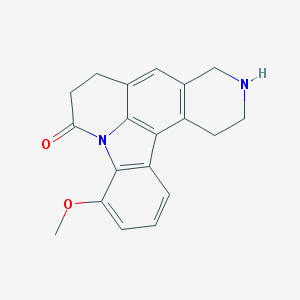 CHEMBL282680 CHEMBL282680 | C19H18N2O2 | 306.365 | 3 / 1 | 2.6 | Yes |
| 3062 | 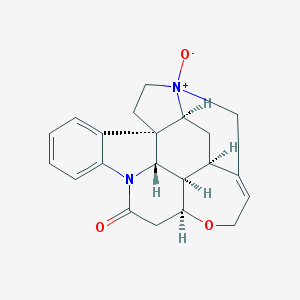 Genostrychnine Genostrychnine | C21H22N2O3 | 350.418 | 3 / 0 | 1.4 | Yes |
| 3137 | 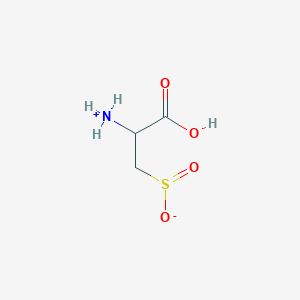 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3656 | 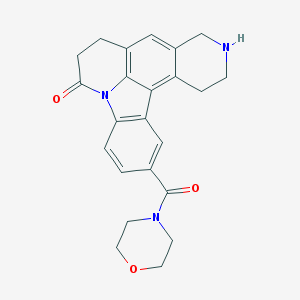 CHEMBL23620 CHEMBL23620 | C23H23N3O3 | 389.455 | 4 / 1 | 1.7 | Yes |
| 3848 | 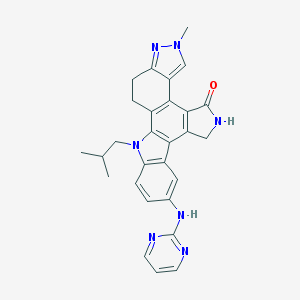 CEP-11981 CEP-11981 | C28H27N7O | 477.572 | 5 / 2 | 3.9 | Yes |
| 4010 | 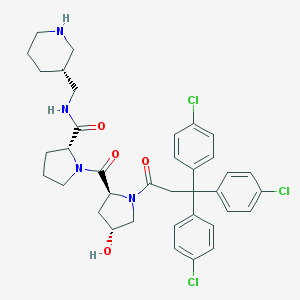 CHEMBL215894 CHEMBL215894 | C37H41Cl3N4O4 | 712.109 | 5 / 3 | 5.9 | No |
| 4317 | 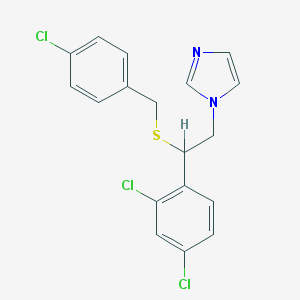 sulconazole sulconazole | C18H15Cl3N2S | 397.742 | 2 / 0 | 6.1 | No |
| 4989 | 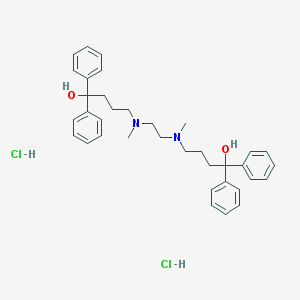 CHEMBL255692 CHEMBL255692 | C36H46Cl2N2O2 | 609.676 | 4 / 4 | N/A | No |
| 5753 | 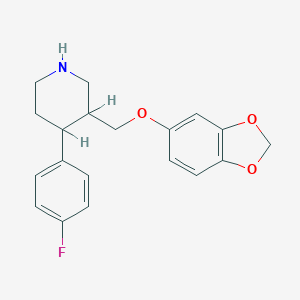 AHOUBRCZNHFOSL-UHFFFAOYSA-N AHOUBRCZNHFOSL-UHFFFAOYSA-N | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5763 | 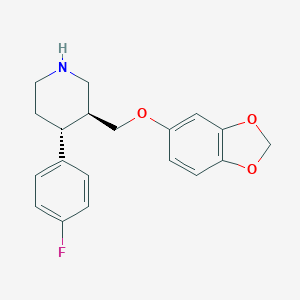 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 553273 |  WIN-51708 WIN-51708 | C29H33N3O | 439.603 | 3 / 1 | 6.5 | No |
| 6331 |  BDBM86295 BDBM86295 | C12H16N3O2+ | 234.279 | 2 / 0 | 0.0 | Yes |
| 6606 |  carbachol carbachol | C6H15ClN2O2 | 182.648 | 3 / 1 | N/A | N/A |
| 6682 |  CHEMBL214029 CHEMBL214029 | C38H43F3N4O4 | 676.781 | 8 / 3 | 4.7 | No |
| 6834 | 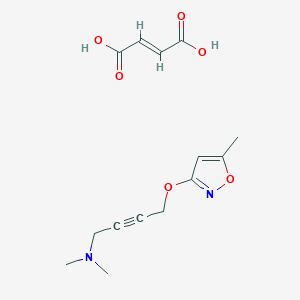 CID 49797317 CID 49797317 | C14H18N2O6 | 310.306 | 8 / 2 | N/A | N/A |
| 463734 | 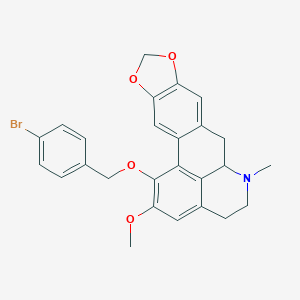 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7189 | 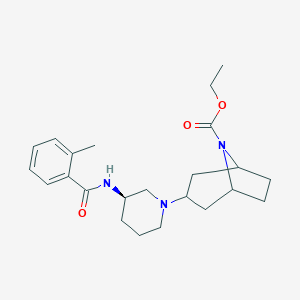 CHEMBL2024331 CHEMBL2024331 | C23H33N3O3 | 399.535 | 4 / 1 | 3.6 | Yes |
| 521664 | 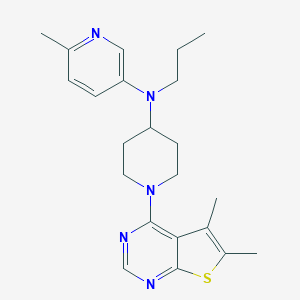 CHEMBL3808947 CHEMBL3808947 | C22H29N5S | 395.569 | 6 / 0 | 5.4 | No |
| 7655 | 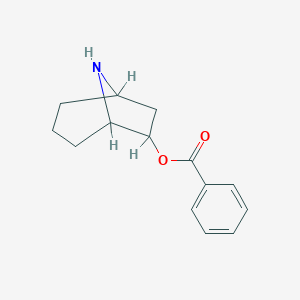 CHEMBL84113 CHEMBL84113 | C14H17NO2 | 231.295 | 3 / 1 | 2.5 | Yes |
| 7778 | 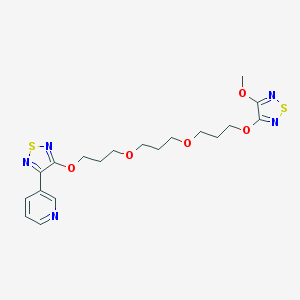 CHEMBL376295 CHEMBL376295 | C19H25N5O5S2 | 467.559 | 12 / 0 | 2.8 | No |
| 521714 | 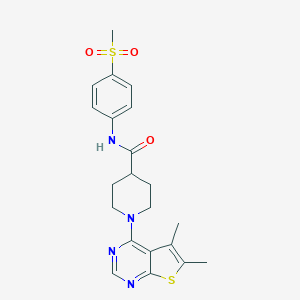 CHEMBL3808592 CHEMBL3808592 | C21H24N4O3S2 | 444.568 | 7 / 1 | 3.3 | Yes |
| 9535 | 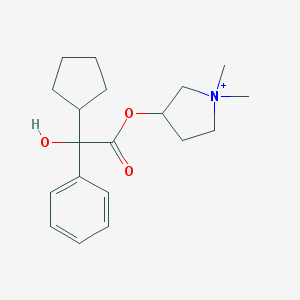 Glycopyrronium Glycopyrronium | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9866 |  AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 9910 |  CHEMBL403773 CHEMBL403773 | C42H52N2O2 | 616.89 | 4 / 2 | 7.9 | No |
| 10172 |  CHEMBL194206 CHEMBL194206 | C23H30FN5O2 | 427.524 | 6 / 1 | 2.7 | Yes |
| 10391 |  N-Allyl Brucine Iodide N-Allyl Brucine Iodide | C26H31N2O4+ | 435.544 | 4 / 0 | 1.6 | Yes |
| 10708 |  CHEMBL213709 CHEMBL213709 | C39H45F3N4O4 | 690.808 | 8 / 2 | 5.1 | No |
| 10721 |  CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 10914 |  CHEMBL169494 CHEMBL169494 | C19H22N2 | 278.399 | 2 / 0 | 2.4 | Yes |
| 11918 |  CHEMBL477821 CHEMBL477821 | C24H29ClN4O | 424.973 | 3 / 1 | 4.1 | Yes |
| 12544 |  CHEMBL593620 CHEMBL593620 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12549 |  CHEMBL593685 CHEMBL593685 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12703 |  CHEMBL1083925 CHEMBL1083925 | C20H22ClFN2O | 360.857 | 3 / 2 | 4.3 | Yes |
| 12819 |  CHEMBL85190 CHEMBL85190 | C15H19NO2 | 245.322 | 3 / 0 | 3.0 | Yes |
| 13309 |  Aclidinium Aclidinium | C26H30NO4S2+ | 484.649 | 6 / 1 | 4.7 | Yes |
| 13781 |  CHEMBL557808 CHEMBL557808 | C19H30INO2S | 463.418 | 4 / 0 | N/A | N/A |
| 14003 |  CHEMBL1922764 CHEMBL1922764 | C19H20ClNO2 | 329.824 | 3 / 0 | 5.7 | No |
| 15153 |  cremastrine cremastrine | C14H25NO3 | 255.358 | 4 / 1 | 2.2 | Yes |
| 15430 | 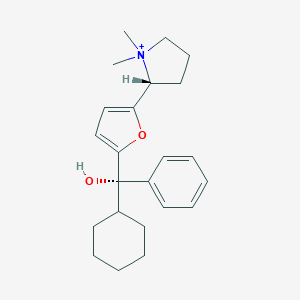 CHEMBL568775 CHEMBL568775 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15434 | 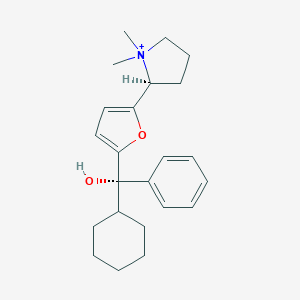 CHEMBL569089 CHEMBL569089 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15438 | 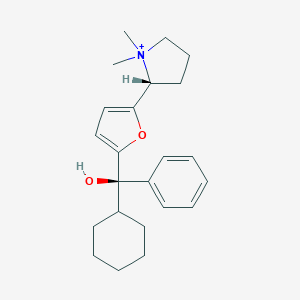 CHEMBL569307 CHEMBL569307 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15445 | 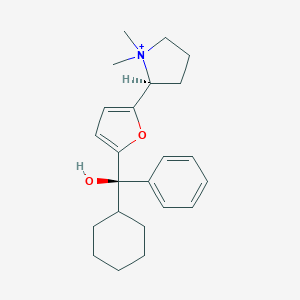 CHEMBL568870 CHEMBL568870 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15451 |  CHEMBL569760 CHEMBL569760 | C23H32NO2+ | 354.514 | 2 / 1 | 4.4 | Yes |
| 15471 |  CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15783 |  CID 44448670 CID 44448670 | C21H28ClNO | 345.911 | 2 / 2 | N/A | N/A |
| 442263 |  CHEMBL3354075 CHEMBL3354075 | C26H42N2O2 | 414.634 | 3 / 1 | 5.7 | No |
| 521953 |  AM-0902 AM-0902 | C17H15ClN6O2 | 370.797 | 6 / 0 | 2.3 | Yes |
| 16574 |  CHEMBL2059306 CHEMBL2059306 | C24H28N4O2 | 404.514 | 3 / 1 | 3.8 | Yes |
| 20177 |  imipramine imipramine | C19H24N2 | 280.415 | 2 / 0 | 4.8 | Yes |
| 20543 |  AQ-RA 741 AQ-RA 741 | C27H37N5O2 | 463.626 | 5 / 1 | 3.1 | Yes |
| 536501 | 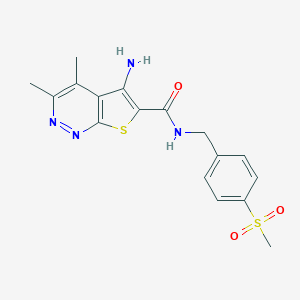 CHEMBL3974708 CHEMBL3974708 | C17H18N4O3S2 | 390.476 | 7 / 2 | 2.2 | Yes |
| 20975 | 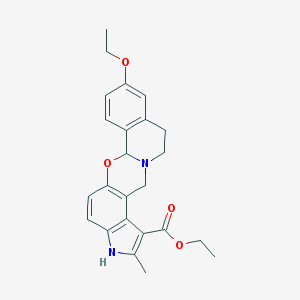 CHEMBL62247 CHEMBL62247 | C24H26N2O4 | 406.482 | 5 / 1 | 4.2 | Yes |
| 21628 | 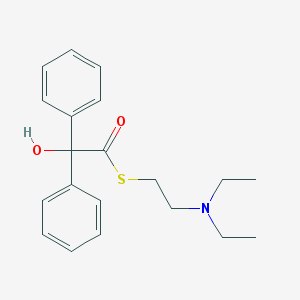 AC1L7RMR AC1L7RMR | C20H25NO2S | 343.485 | 4 / 1 | 4.0 | Yes |
| 22069 | 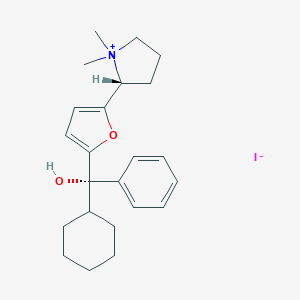 CHEMBL568775 CHEMBL568775 | C23H32INO2 | 481.418 | 3 / 1 | N/A | N/A |
| 22074 |  CHEMBL569760 CHEMBL569760 | C23H32INO2 | 481.418 | 3 / 1 | N/A | N/A |
| 22076 |  CHEMBL569089 CHEMBL569089 | C23H32INO2 | 481.418 | 3 / 1 | N/A | N/A |
| 22080 |  CHEMBL568870 CHEMBL568870 | C23H32INO2 | 481.418 | 3 / 1 | N/A | N/A |
| 22089 |  CHEMBL569307 CHEMBL569307 | C23H32INO2 | 481.418 | 3 / 1 | N/A | N/A |
| 522128 | 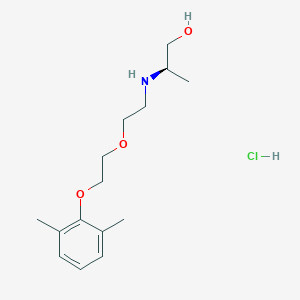 CHEMBL3775436 CHEMBL3775436 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 522181 | 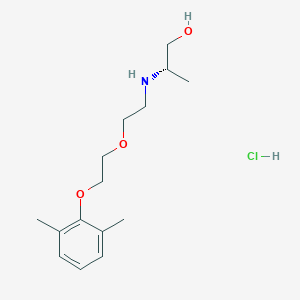 CHEMBL3775769 CHEMBL3775769 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 22179 | 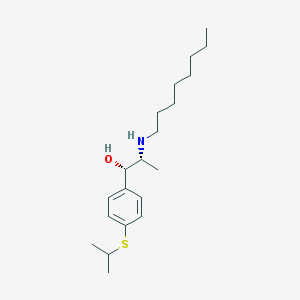 suloctidil suloctidil | C20H35NOS | 337.566 | 3 / 2 | 5.8 | No |
| 22243 | 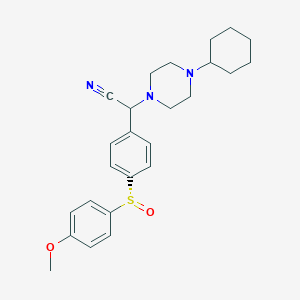 CHEMBL2111540 CHEMBL2111540 | C25H31N3O2S | 437.602 | 6 / 0 | 4.1 | Yes |
| 22245 |  CHEMBL2114066 CHEMBL2114066 | C25H31N3O2S | 437.602 | 6 / 0 | 4.1 | Yes |
| 22550 |  CHEMBL283036 CHEMBL283036 | C19H19F3N4 | 360.384 | 6 / 1 | 3.6 | Yes |
| 22873 |  tropicamide tropicamide | C17H20N2O2 | 284.359 | 3 / 1 | 1.5 | Yes |
| 23578 |  CHEMBL378872 CHEMBL378872 | C37H41Cl3N4O4 | 712.109 | 5 / 3 | 5.9 | No |
| 24595 |  CHEMBL262198 CHEMBL262198 | C24H27NO6 | 425.481 | 7 / 2 | N/A | N/A |
| 24636 |  CHEMBL540316 CHEMBL540316 | C23H28ClNO2 | 385.932 | 3 / 1 | N/A | N/A |
| 25533 |  CHEMBL2313391 CHEMBL2313391 | C18H16ClN3O3S | 389.854 | 6 / 2 | 4.4 | Yes |
| 25801 |  CHEMBL2042406 CHEMBL2042406 | C20H26INO2 | 439.337 | 3 / 0 | N/A | N/A |
| 25806 |  CHEMBL2042405 CHEMBL2042405 | C20H26INO2 | 439.337 | 3 / 0 | N/A | N/A |
| 25808 |  CHEMBL2042404 CHEMBL2042404 | C20H26INO2 | 439.337 | 3 / 0 | N/A | N/A |
| 522354 |  CHEMBL1314270 CHEMBL1314270 | C20H20Cl2N4OS | 435.367 | 5 / 1 | 5.4 | No |
| 26382 |  CHEMBL105122 CHEMBL105122 | C23H24N2O3S | 408.516 | 5 / 1 | 4.4 | Yes |
| 26439 | 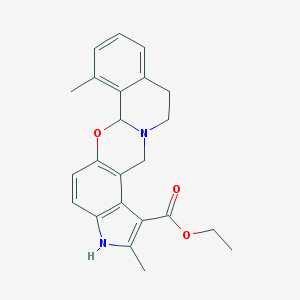 CHEMBL304036 CHEMBL304036 | C23H24N2O3 | 376.456 | 4 / 1 | 4.3 | Yes |
| 26453 | 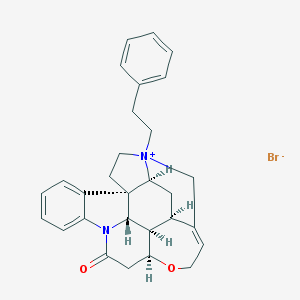 CHEMBL1202234 CHEMBL1202234 | C29H31BrN2O2 | 519.483 | 3 / 0 | N/A | No |
| 26526 | 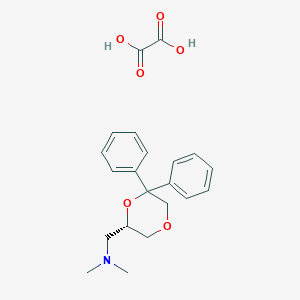 CHEMBL2042553 CHEMBL2042553 | C21H25NO6 | 387.432 | 7 / 2 | N/A | N/A |
| 26528 | 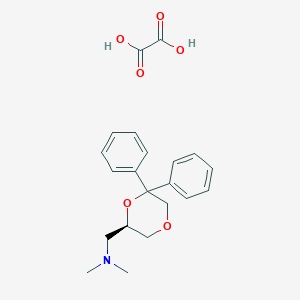 CHEMBL2042554 CHEMBL2042554 | C21H25NO6 | 387.432 | 7 / 2 | N/A | N/A |
| 26535 | 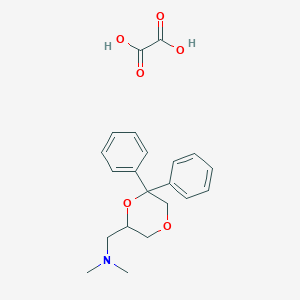 CHEMBL2042552 CHEMBL2042552 | C21H25NO6 | 387.432 | 7 / 2 | N/A | N/A |
| 26938 | 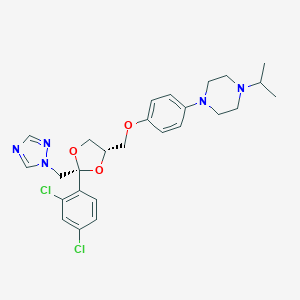 terconazole terconazole | C26H31Cl2N5O3 | 532.466 | 7 / 0 | 4.8 | No |
| 27203 | 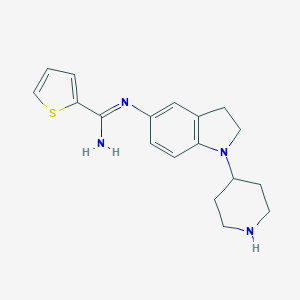 CHEMBL2203713 CHEMBL2203713 | C18H22N4S | 326.462 | 4 / 2 | 2.9 | Yes |
| 27267 | 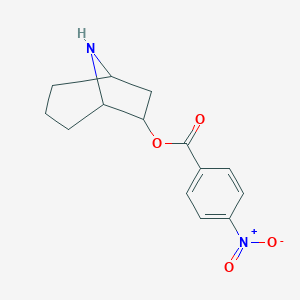 CHEMBL315428 CHEMBL315428 | C14H16N2O4 | 276.292 | 5 / 1 | 2.3 | Yes |
| 27495 |  CHEMBL2432063 CHEMBL2432063 | C19H29N | 271.448 | 1 / 0 | 4.7 | Yes |
| 28360 |  CHEMBL593685 CHEMBL593685 | C11H24NO2+ | 202.318 | 2 / 0 | 0.9 | Yes |
| 28362 |  CHEMBL593620 CHEMBL593620 | C11H24NO2+ | 202.318 | 2 / 0 | 0.9 | Yes |
| 28524 |  CHEMBL197796 CHEMBL197796 | C24H33N5O2S | 455.621 | 6 / 1 | 3.1 | Yes |
| 28620 |  CHEMBL352779 CHEMBL352779 | C20H23N3O | 321.424 | 3 / 1 | 2.2 | Yes |
| 28871 |  CHEMBL280340 CHEMBL280340 | C22H24N2O3 | 364.445 | 4 / 1 | 3.4 | Yes |
| 28941 |  CHEMBL2407171 CHEMBL2407171 | C20H28N4O4 | 388.468 | 6 / 1 | 1.7 | Yes |
| 29603 |  CHEMBL2336054 CHEMBL2336054 | C21H29N3O3 | 371.481 | 4 / 0 | 2.2 | Yes |
| 29609 | 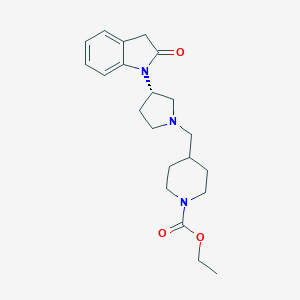 CHEMBL2336055 CHEMBL2336055 | C21H29N3O3 | 371.481 | 4 / 0 | 2.2 | Yes |
| 31077 | 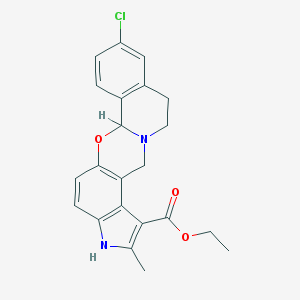 CHEMBL105674 CHEMBL105674 | C22H21ClN2O3 | 396.871 | 4 / 1 | 4.5 | Yes |
| 31813 | 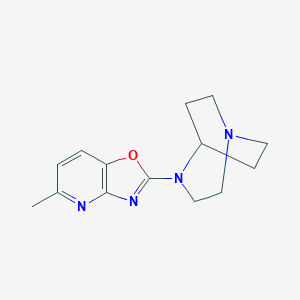 CP-810123 CP-810123 | C14H18N4O | 258.325 | 5 / 0 | 2.2 | Yes |
| 31976 | 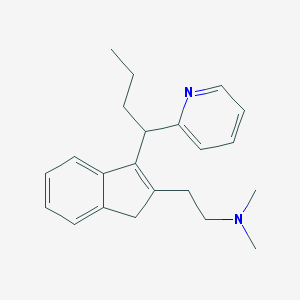 CHEMBL170088 CHEMBL170088 | C22H28N2 | 320.48 | 2 / 0 | 3.6 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417