

You can:
| Name | Muscarinic acetylcholine receptor M2 |
|---|---|
| Species | Rattus norvegicus (Rat) |
| Gene | Chrm2 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 2 AChR M2 Chrm-2 M2 muscarinic acetylcholine receptor [ Show all ] |
| Disease | N/A for non-human GPCRs |
| Length | 466 |
| Amino acid sequence | MNNSTNSSNNGLAITSPYKTFEVVFIVLVAGSLSLVTIIGNILVMVSIKVNRHLQTVNNYFLFSLACADLIIGVFSMNLYTLYTVIGYWPLGPVVCDLWLALDYVVSNASVMNLLIISFDRYFCVTKPLTYPVKRTTKMAGMMIAAAWVLSFILWAPAILFWQFIVGVRTVEDGECYIQFFSNAAVTFGTAIAAFYLPVIIMTVLYWHISRASKSRIKKEKKEPVANQDPVSPSLVQGRIVKPNNNNMPGGDGGLEHNKIQNGKAPRDGVTENCVQGEEKESSNDSTSVSAVASNMRDDEITQDENTVSTSLGHSRDDNSKQTCIKIVTKAQKGDVCTPTSTTVELVGSSGQNGDEKQNIVARKIVKMTKQPAKKKPPPSREKKVTRTILAILLAFIITWAPYNVMVLINTFCAPCIPNTVWTIGYWLCYINSTINPACYALCNATFKKTFKHLLMCHYKNIGATR |
| UniProt | P10980 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL309 |
| IUPHAR | 14 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 93 |  Eticlopride Eticlopride | C17H25ClN2O3 | 340.848 | 4 / 2 | 3.1 | Yes |
| 1331 |  CHEMBL68292 CHEMBL68292 | C29H40N6O3 | 520.678 | 6 / 2 | 2.5 | No |
| 1416 |  1-(4-methylpiperazin-1-yl)-4-phenylphthalazine 1-(4-methylpiperazin-1-yl)-4-phenylphthalazine | C19H20N4 | 304.397 | 4 / 0 | 3.0 | Yes |
| 2980 |  CHEMBL553066 CHEMBL553066 | C17H23ClN2O3 | 338.832 | 4 / 3 | N/A | N/A |
| 3092 | 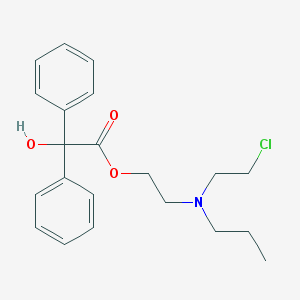 PROPYLBENZILYLCHOLINE MUSTARD PROPYLBENZILYLCHOLINE MUSTARD | C21H26ClNO3 | 375.893 | 4 / 1 | 4.2 | Yes |
| 3683 | 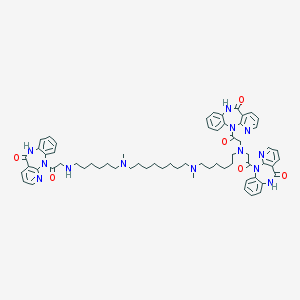 CHEMBL1202000 CHEMBL1202000 | C64H77N13O6 | 1124.41 | 13 / 4 | 7.7 | No |
| 3960 | 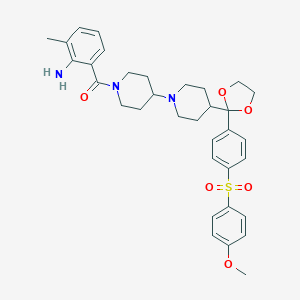 CHEMBL332703 CHEMBL332703 | C34H41N3O6S | 619.777 | 8 / 1 | 4.6 | No |
| 4962 | 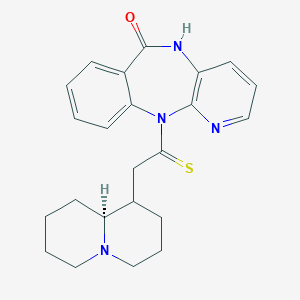 CHEMBL103189 CHEMBL103189 | C23H26N4OS | 406.548 | 4 / 1 | 2.4 | Yes |
| 5756 | 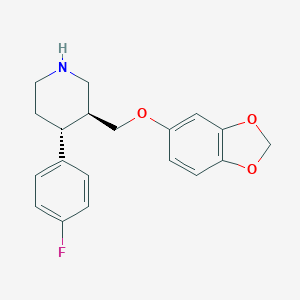 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 6384 | 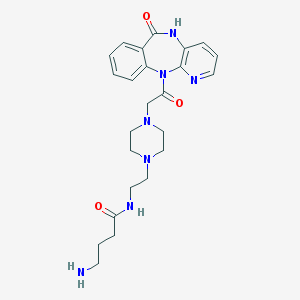 CHEMBL302604 CHEMBL302604 | C24H31N7O3 | 465.558 | 7 / 3 | -1.2 | Yes |
| 6614 | 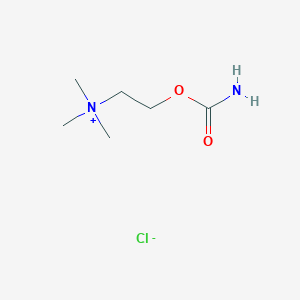 carbachol carbachol | C6H15ClN2O2 | 182.648 | 3 / 1 | N/A | N/A |
| 7634 | 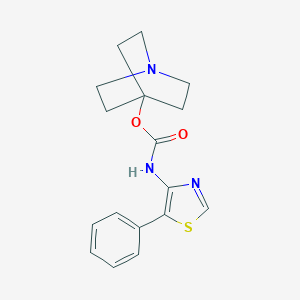 CHEMBL3298591 CHEMBL3298591 | C17H19N3O2S | 329.418 | 5 / 1 | 3.1 | Yes |
| 8155 |  CHEMBL70154 CHEMBL70154 | C30H42N6O3 | 534.705 | 6 / 2 | 3.0 | No |
| 8680 |  CHEMBL540013 CHEMBL540013 | C26H33NO2 | 391.555 | 3 / 0 | 5.8 | No |
| 12281 |  CHEMBL540013 CHEMBL540013 | C26H34ClNO2 | 428.013 | 3 / 1 | N/A | N/A |
| 13777 |  CHEMBL304109 CHEMBL304109 | C33H39N7O5 | 613.719 | 8 / 3 | 1.6 | No |
| 15159 | 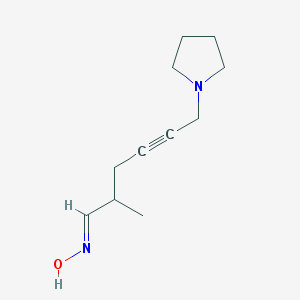 D0U2IL D0U2IL | C11H18N2O | 194.278 | 3 / 1 | 1.5 | Yes |
| 15418 | 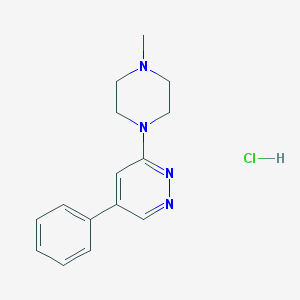 CHEMBL540310 CHEMBL540310 | C15H19ClN4 | 290.795 | 4 / 1 | N/A | N/A |
| 16194 | 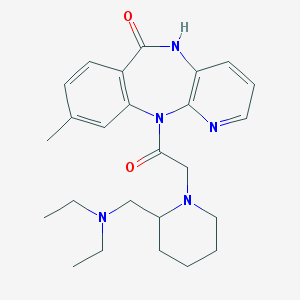 CHEMBL294919 CHEMBL294919 | C25H33N5O2 | 435.572 | 5 / 1 | 2.3 | Yes |
| 16789 | 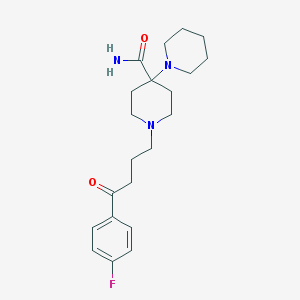 Pipamperone Pipamperone | C21H30FN3O2 | 375.488 | 5 / 1 | 2.0 | Yes |
| 17057 | 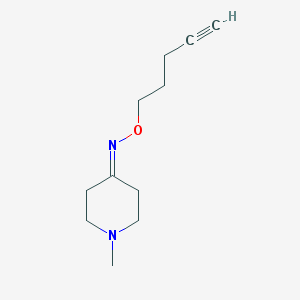 CHEMBL321842 CHEMBL321842 | C11H18N2O | 194.278 | 3 / 0 | 1.2 | Yes |
| 17222 | 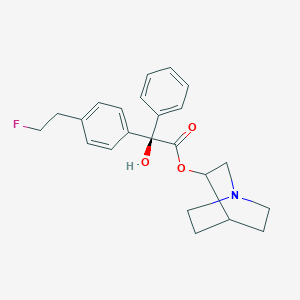 CHEMBL49576 CHEMBL49576 | C23H26FNO3 | 383.463 | 5 / 1 | 3.9 | Yes |
| 17223 | 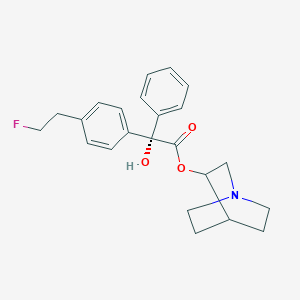 CHEMBL296633 CHEMBL296633 | C23H26FNO3 | 383.463 | 5 / 1 | 3.9 | Yes |
| 19929 | 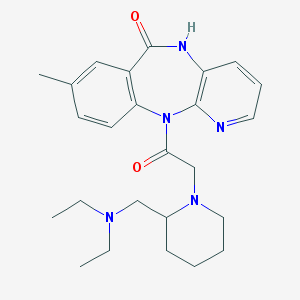 CHEMBL297224 CHEMBL297224 | C25H33N5O2 | 435.572 | 5 / 1 | 2.3 | Yes |
| 20176 | 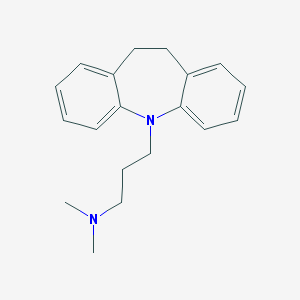 imipramine imipramine | C19H24N2 | 280.415 | 2 / 0 | 4.8 | Yes |
| 20540 | 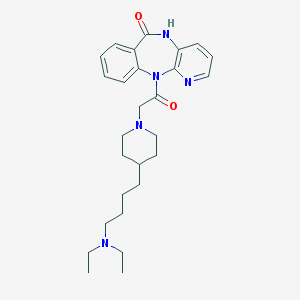 AQ-RA 741 AQ-RA 741 | C27H37N5O2 | 463.626 | 5 / 1 | 3.1 | Yes |
| 20819 | 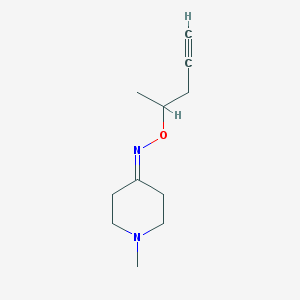 CHEMBL320121 CHEMBL320121 | C11H18N2O | 194.278 | 3 / 0 | 1.3 | Yes |
| 21403 | 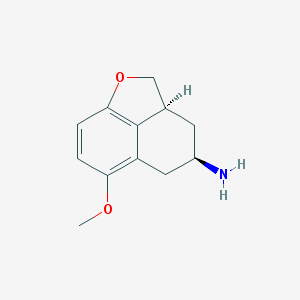 CHEMBL68240 CHEMBL68240 | C12H15NO2 | 205.257 | 3 / 1 | 1.2 | Yes |
| 22139 |  CHEMBL265521 CHEMBL265521 | C13H20N2O | 220.316 | 3 / 0 | 1.9 | Yes |
| 22228 |  CHEMBL296405 CHEMBL296405 | C25H33N5O2 | 435.572 | 5 / 1 | 2.3 | Yes |
| 22821 |  CHEMBL68771 CHEMBL68771 | C20H24N6O2 | 380.452 | 6 / 2 | -0.8 | Yes |
| 22871 |  tropicamide tropicamide | C17H20N2O2 | 284.359 | 3 / 1 | 1.5 | Yes |
| 23196 |  CHEMBL285803 CHEMBL285803 | C8H12INO2S | 313.153 | 3 / 0 | N/A | N/A |
| 23359 |  sulpiride sulpiride | C15H23N3O4S | 341.426 | 6 / 2 | 0.6 | Yes |
| 25946 |  CHEMBL43601 CHEMBL43601 | C30H42N4O2 | 490.692 | 4 / 1 | 5.6 | No |
| 27437 |  CHEMBL305403 CHEMBL305403 | C28H31N7O3 | 513.602 | 7 / 3 | -0.1 | No |
| 28983 | 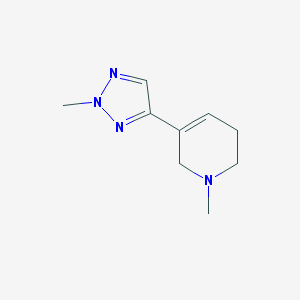 CHEMBL129886 CHEMBL129886 | C9H14N4 | 178.239 | 3 / 0 | 0.5 | Yes |
| 29632 | 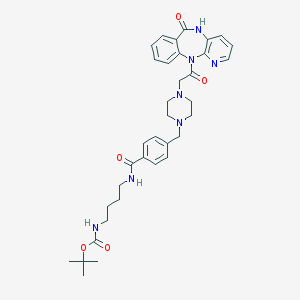 CHEMBL69458 CHEMBL69458 | C35H43N7O5 | 641.773 | 8 / 3 | 2.3 | No |
| 29797 | 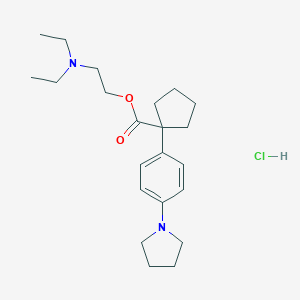 CHEMBL542914 CHEMBL542914 | C22H35ClN2O2 | 394.984 | 4 / 1 | N/A | N/A |
| 29981 | 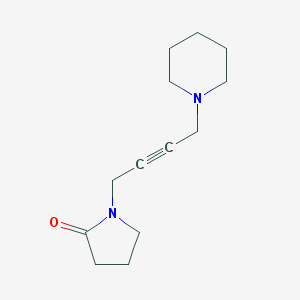 CHEMBL118240 CHEMBL118240 | C13H20N2O | 220.316 | 2 / 0 | 0.7 | Yes |
| 30614 |  CHEMBL116777 CHEMBL116777 | C34H40N2O6S | 604.762 | 7 / 0 | 4.8 | No |
| 30625 |  CHEMBL160755 CHEMBL160755 | C24H39NO3 | 389.58 | 4 / 0 | 5.4 | No |
| 30854 |  CHEMBL312200 CHEMBL312200 | C11H20N4O2 | 240.307 | 4 / 2 | -0.3 | Yes |
| 30961 |  CHEMBL303678 CHEMBL303678 | C20H21N5O3 | 379.42 | 5 / 1 | -0.4 | Yes |
| 31635 |  CHEMBL25649 CHEMBL25649 | C9H15N2O2+ | 183.231 | 3 / 0 | 0.4 | Yes |
| 32036 |  CHEMBL159189 CHEMBL159189 | C26H43NO4 | 433.633 | 5 / 0 | 5.5 | No |
| 32431 |  CHEMBL3298589 CHEMBL3298589 | C18H21ClN2O2S | 364.888 | 4 / 2 | N/A | N/A |
| 32433 |  CHEMBL3298590 CHEMBL3298590 | C18H21ClN2O2S | 364.888 | 4 / 2 | N/A | N/A |
| 32496 |  Triazoledione Triazoledione | C23H28ClN5O3 | 457.959 | 5 / 1 | 3.6 | Yes |
| 32926 |  CHEMBL119294 CHEMBL119294 | C34H38N2O7S | 618.745 | 8 / 0 | 4.6 | No |
| 33721 |  CHEMBL304112 CHEMBL304112 | C30H30N6O4 | 538.608 | 7 / 1 | 1.5 | No |
| 35019 |  CHEMBL326990 CHEMBL326990 | C34H41N3O6S | 619.777 | 8 / 1 | 4.9 | No |
| 35192 | 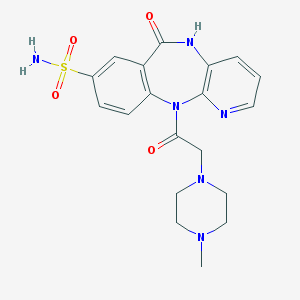 CHEMBL304599 CHEMBL304599 | C19H22N6O4S | 430.483 | 8 / 2 | -1.3 | Yes |
| 35602 | 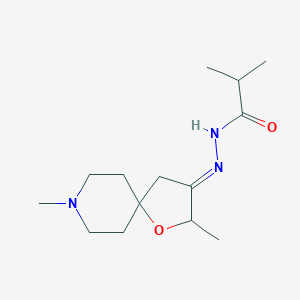 CHEMBL85729 CHEMBL85729 | C14H25N3O2 | 267.373 | 4 / 1 | 1.3 | Yes |
| 36745 | 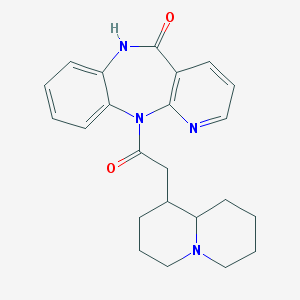 CHEMBL103529 CHEMBL103529 | C23H26N4O2 | 390.487 | 4 / 1 | 2.6 | Yes |
| 36807 | 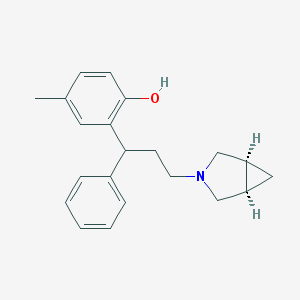 CHEMBL190815 CHEMBL190815 | C21H25NO | 307.437 | 2 / 1 | 4.5 | Yes |
| 37363 |  Oxotremorine M Oxotremorine M | C11H19N2O+ | 195.286 | 1 / 0 | -0.1 | Yes |
| 37694 |  CHEMBL1916225 CHEMBL1916225 | C21H30FN3O3 | 391.487 | 5 / 1 | 3.1 | Yes |
| 37923 |  CHEMBL360491 CHEMBL360491 | C24H26N2O2 | 374.484 | 3 / 0 | 3.2 | Yes |
| 37954 |  CHEMBL68709 CHEMBL68709 | C25H23N5O3 | 441.491 | 5 / 1 | 1.2 | Yes |
| 38600 | 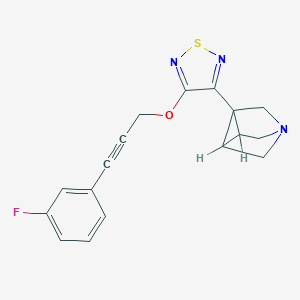 CHEMBL304247 CHEMBL304247 | C17H14FN3OS | 327.377 | 6 / 0 | 2.6 | Yes |
| 39212 | 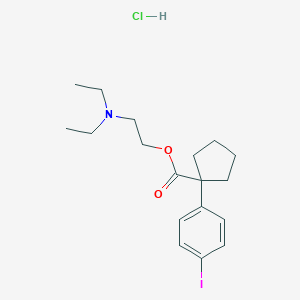 CHEMBL544107 CHEMBL544107 | C18H27ClINO2 | 451.773 | 3 / 1 | N/A | N/A |
| 553442 | 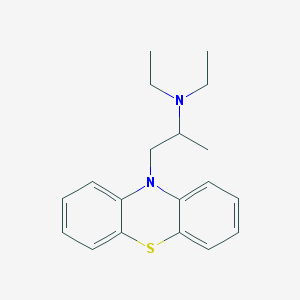 ethopropazine ethopropazine | C19H24N2S | 312.475 | 3 / 0 | 4.8 | Yes |
| 39780 | 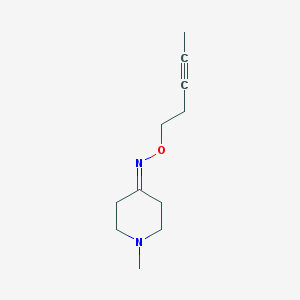 CHEMBL322293 CHEMBL322293 | C11H18N2O | 194.278 | 3 / 0 | 1.4 | Yes |
| 40151 | 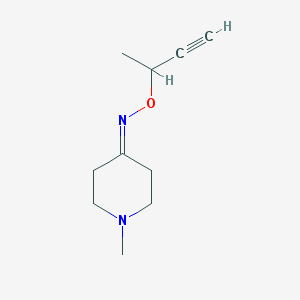 CHEMBL109120 CHEMBL109120 | C10H16N2O | 180.251 | 3 / 0 | 0.8 | Yes |
| 40935 | 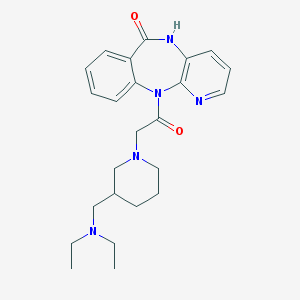 CHEMBL295214 CHEMBL295214 | C24H31N5O2 | 421.545 | 5 / 1 | 1.8 | Yes |
| 42084 | 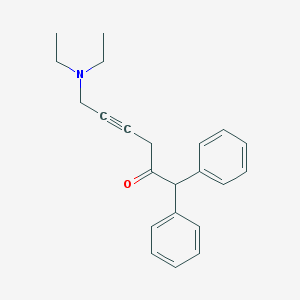 DABDMA DABDMA | C22H25NO | 319.448 | 2 / 0 | 4.2 | Yes |
| 42745 | 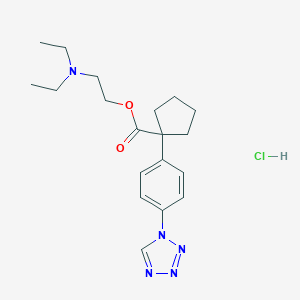 CHEMBL542915 CHEMBL542915 | C19H28ClN5O2 | 393.916 | 6 / 1 | N/A | N/A |
| 43549 |  fluvoxamine fluvoxamine | C15H21F3N2O2 | 318.34 | 7 / 1 | 2.6 | Yes |
| 43682 |  CHEMBL278618 CHEMBL278618 | C8H12NO2S+ | 186.249 | 3 / 0 | 1.1 | Yes |
| 43981 |  DMCAE DMCAE | C7H16N2O2 | 160.217 | 3 / 0 | 0.6 | Yes |
| 44051 |  CHEMBL553865 CHEMBL553865 | C12H21ClN2O2S | 292.822 | 4 / 1 | N/A | N/A |
| 44164 |  VU 0238429 VU 0238429 | C17H12F3NO4 | 351.281 | 7 / 0 | 3.5 | Yes |
| 44541 |  CHEMBL276031 CHEMBL276031 | C19H26N2O | 298.43 | 2 / 1 | 3.9 | Yes |
| 45111 |  CHEMBL62429 CHEMBL62429 | C19H30N2O3 | 334.46 | 4 / 2 | 3.9 | Yes |
| 45151 |  CHEMBL445106 CHEMBL445106 | C12H14BrNO2 | 284.153 | 3 / 1 | 1.9 | Yes |
| 45939 | 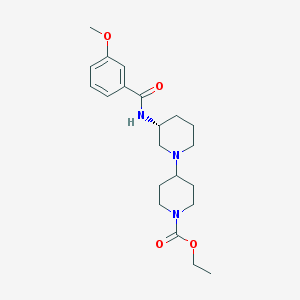 CHEMBL1916228 CHEMBL1916228 | C21H31N3O4 | 389.496 | 5 / 1 | 2.6 | Yes |
| 46233 | 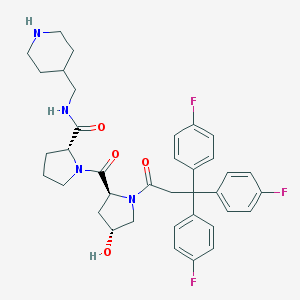 CHEMBL215180 CHEMBL215180 | C37H41F3N4O4 | 662.754 | 8 / 3 | 4.3 | No |
| 47329 | 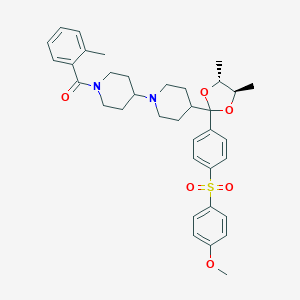 CHEMBL115642 CHEMBL115642 | C36H44N2O6S | 632.816 | 7 / 0 | 5.6 | No |
| 47375 | 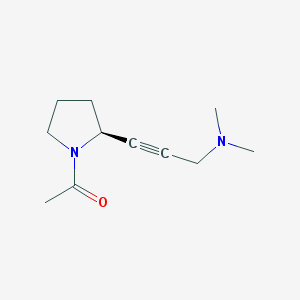 CHEMBL108411 CHEMBL108411 | C11H18N2O | 194.278 | 2 / 0 | 0.3 | Yes |
| 47377 | 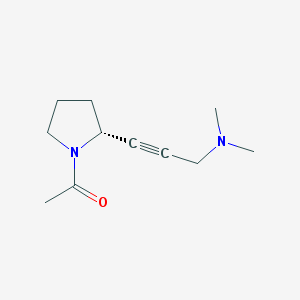 CHEMBL2111698 CHEMBL2111698 | C11H18N2O | 194.278 | 2 / 0 | 0.3 | Yes |
| 47437 | 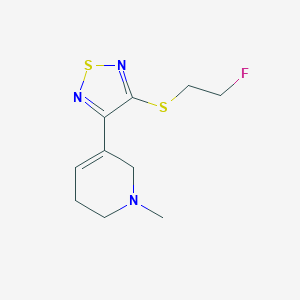 CHEMBL2112938 CHEMBL2112938 | C10H14FN3S2 | 259.361 | 6 / 0 | 1.8 | Yes |
| 47451 | 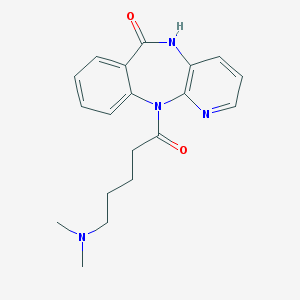 CHEMBL279135 CHEMBL279135 | C19H22N4O2 | 338.411 | 4 / 1 | 0.9 | Yes |
| 47740 | 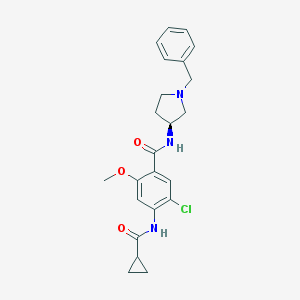 CHEMBL328866 CHEMBL328866 | C23H26ClN3O3 | 427.929 | 4 / 2 | 3.2 | Yes |
| 48250 | 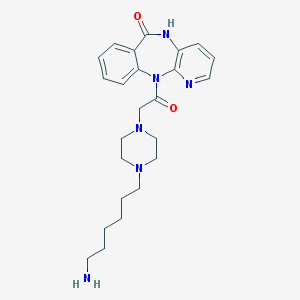 CHEMBL305197 CHEMBL305197 | C24H32N6O2 | 436.56 | 6 / 2 | 0.6 | Yes |
| 48647 | 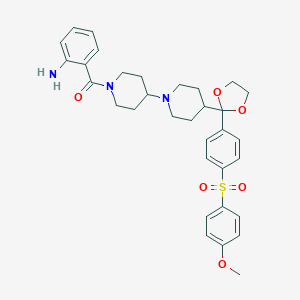 CHEMBL117797 CHEMBL117797 | C33H39N3O6S | 605.75 | 8 / 1 | 4.3 | No |
| 48873 | 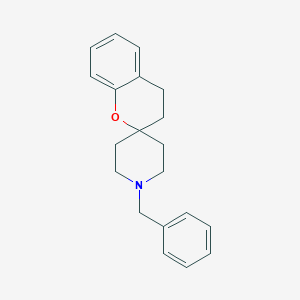 CHEMBL38634 CHEMBL38634 | C20H23NO | 293.41 | 2 / 0 | 4.2 | Yes |
| 48913 | 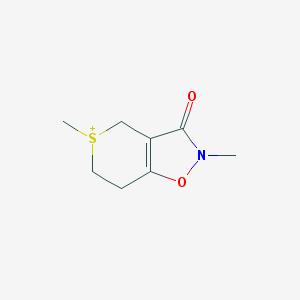 CHEMBL285803 CHEMBL285803 | C8H12NO2S+ | 186.249 | 2 / 0 | 0.4 | Yes |
| 49673 |  CHEMBL194870 CHEMBL194870 | C23H22F2N2O2 | 396.438 | 5 / 2 | 2.8 | Yes |
| 50432 |  CHEMBL1202002 CHEMBL1202002 | C78H86N16O8 | 1375.65 | 16 / 4 | 9.0 | No |
| 51058 |  dicyclomine dicyclomine | C19H35NO2 | 309.494 | 3 / 0 | 5.5 | No |
| 51900 |  CHEMBL112244 CHEMBL112244 | C36H59N7O2 | 621.915 | 7 / 3 | 4.3 | No |
| 52295 |  TBPB TBPB | C25H32N4O | 404.558 | 3 / 1 | 4.4 | Yes |
| 52461 |  CHEMBL300660 CHEMBL300660 | C25H33N5O2 | 435.572 | 5 / 1 | 2.5 | Yes |
| 52744 |  McN-A-343 McN-A-343 | C14H18Cl2N2O2 | 317.21 | 3 / 1 | N/A | N/A |
| 52938 |  CHEMBL296365 CHEMBL296365 | C22H27N5O2 | 393.491 | 5 / 2 | 1.1 | Yes |
| 53265 | 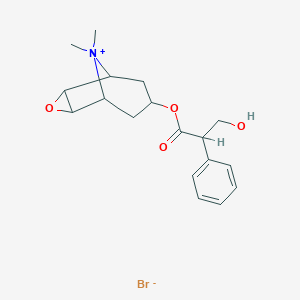 (-)-Scopolamine methyl bromide (-)-Scopolamine methyl bromide | C18H24BrNO4 | 398.297 | 5 / 1 | N/A | N/A |
| 53448 | 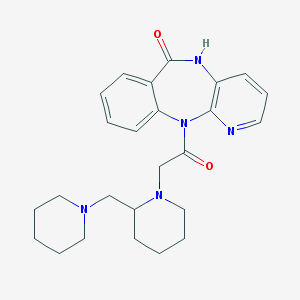 CHEMBL45983 CHEMBL45983 | C25H31N5O2 | 433.556 | 5 / 1 | 2.1 | Yes |
| 53909 | 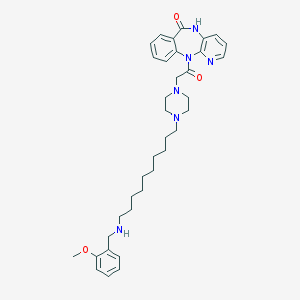 CHEMBL68446 CHEMBL68446 | C36H48N6O3 | 612.819 | 7 / 2 | 4.7 | No |
| 55054 | 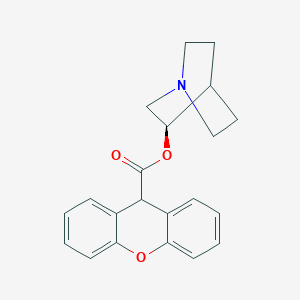 CHEMBL549577 CHEMBL549577 | C21H21NO3 | 335.403 | 4 / 0 | 3.2 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417