

You can:
| Name | Muscarinic acetylcholine receptor M1 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CHRM1 |
| Synonym | cholinergic receptor cholinergic receptor, muscarinic 1 cholinergic receptor, muscarinic 1, CNS Chrm-1 M1 receptor [ Show all ] |
| Disease | Functional bowel syndrome; Irritable bowel syndrome Glaucoma Peptic ulcer Parkinsonism; Extrapyramidal disorders secondary to neuroleptic drug therapy Visceral spasms [ Show all ] |
| Length | 460 |
| Amino acid sequence | MNTSAPPAVSPNITVLAPGKGPWQVAFIGITTGLLSLATVTGNLLVLISFKVNTELKTVNNYFLLSLACADLIIGTFSMNLYTTYLLMGHWALGTLACDLWLALDYVASNASVMNLLLISFDRYFSVTRPLSYRAKRTPRRAALMIGLAWLVSFVLWAPAILFWQYLVGERTVLAGQCYIQFLSQPIITFGTAMAAFYLPVTVMCTLYWRIYRETENRARELAALQGSETPGKGGGSSSSSERSQPGAEGSPETPPGRCCRCCRAPRLLQAYSWKEEEEEDEGSMESLTSSEGEEPGSEVVIKMPMVDPEAQAPTKQPPRSSPNTVKRPTKKGRDRAGKGQKPRGKEQLAKRKTFSLVKEKKAARTLSAILLAFILTWTPYNIMVLVSTFCKDCVPETLWELGYWLCYVNSTINPMCYALCNKAFRDTFRLLLLCRWDKRRWRKIPKRPGSVHRTPSRQC |
| UniProt | P11229 |
| Protein Data Bank | 5cxv |
| GPCR-HGmod model | P11229 |
| 3D structure model | This structure is from PDB ID 5cxv. |
| BioLiP | BL0339262, BL0339261, BL0339263 |
| Therapeutic Target Database | T28893 |
| ChEMBL | CHEMBL216 |
| IUPHAR | 13 |
| DrugBank | BE0000092 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 131 | 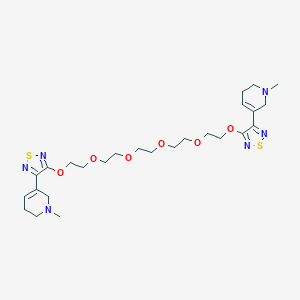 CHEMBL140142 CHEMBL140142 | C26H40N6O6S2 | 596.762 | 14 / 0 | 1.0 | No |
| 156 | 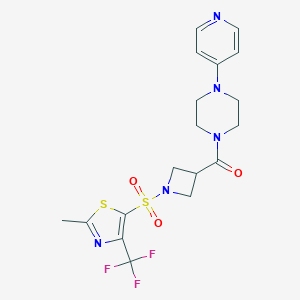 CHEMBL2057501 CHEMBL2057501 | C18H20F3N5O3S2 | 475.505 | 11 / 0 | 1.5 | No |
| 219 | 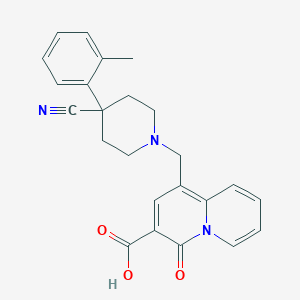 CHEMBL1800884 CHEMBL1800884 | C24H23N3O3 | 401.466 | 5 / 1 | 0.8 | Yes |
| 441678 | 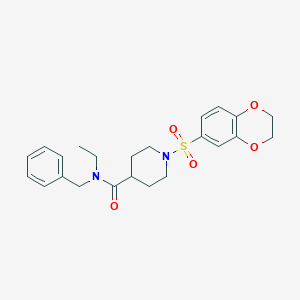 CHEMBL1602658 CHEMBL1602658 | C23H28N2O5S | 444.546 | 6 / 0 | 2.6 | Yes |
| 301 | 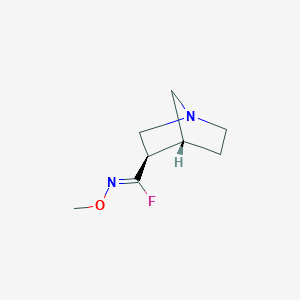 CHEMBL341379 CHEMBL341379 | C8H13FN2O | 172.203 | 4 / 0 | 1.0 | Yes |
| 535920 | 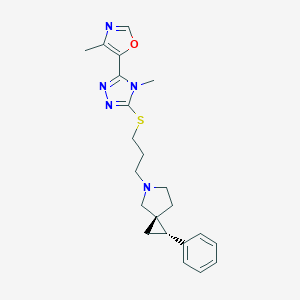 CHEMBL3983937 CHEMBL3983937 | C22H27N5OS | 409.552 | 6 / 0 | 3.3 | Yes |
| 523 | 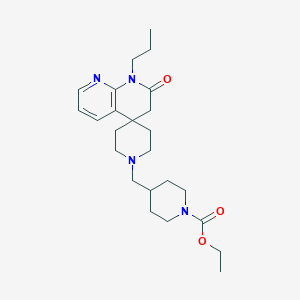 CHEMBL2407175 CHEMBL2407175 | C24H36N4O3 | 428.577 | 5 / 0 | 2.7 | Yes |
| 736 | 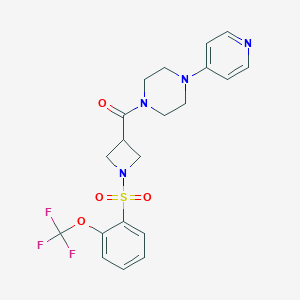 CHEMBL2058899 CHEMBL2058899 | C20H21F3N4O4S | 470.467 | 10 / 0 | 2.0 | Yes |
| 763 | 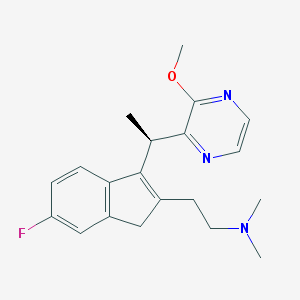 CHEMBL1090526 CHEMBL1090526 | C20H24FN3O | 341.43 | 5 / 0 | 2.7 | Yes |
| 812 | 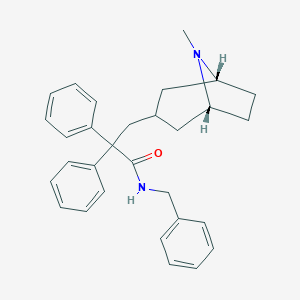 CHEMBL563305 CHEMBL563305 | C30H34N2O | 438.615 | 2 / 1 | 5.9 | No |
| 1004 | 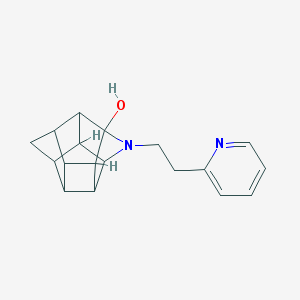 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1054 | 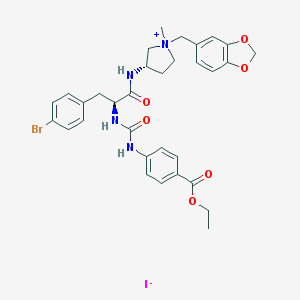 CHEMBL466271 CHEMBL466271 | C32H36BrIN4O6 | 779.47 | 7 / 3 | N/A | No |
| 1187 | 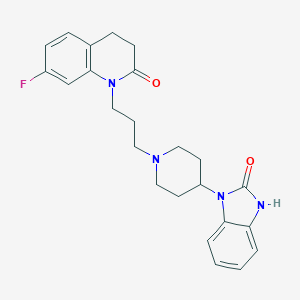 CHEMBL2059319 CHEMBL2059319 | C24H27FN4O2 | 422.504 | 4 / 1 | 3.9 | Yes |
| 1277 | 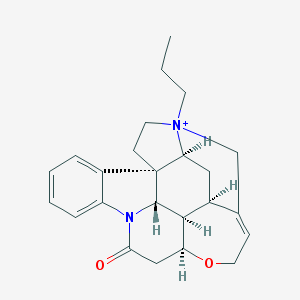 CHEMBL141736 CHEMBL141736 | C24H29N2O2+ | 377.508 | 2 / 0 | 2.8 | Yes |
| 1940 | 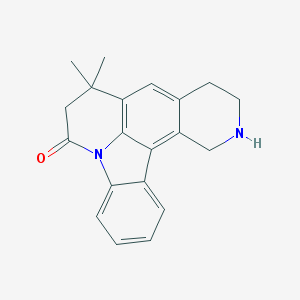 CHEMBL282874 CHEMBL282874 | C20H20N2O | 304.393 | 2 / 1 | 3.5 | Yes |
| 2264 | 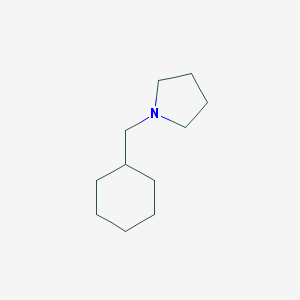 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2321 |  CHEMBL366819 CHEMBL366819 | C22H37NO2 | 347.543 | 3 / 0 | 5.4 | No |
| 2908 |  CHEMBL282680 CHEMBL282680 | C19H18N2O2 | 306.365 | 3 / 1 | 2.6 | Yes |
| 3063 |  Genostrychnine Genostrychnine | C21H22N2O3 | 350.418 | 3 / 0 | 1.4 | Yes |
| 3135 |  AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3178 | 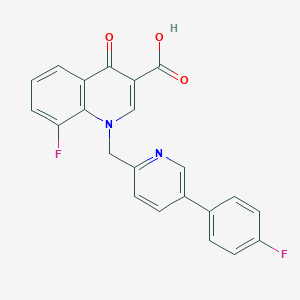 CHEMBL600482 CHEMBL600482 | C22H14F2N2O3 | 392.362 | 7 / 1 | 4.3 | Yes |
| 557378 | 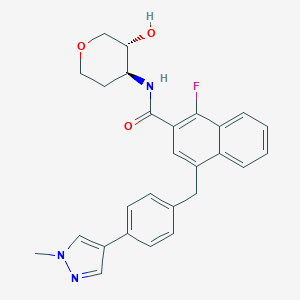 SCHEMBL18211801 SCHEMBL18211801 | C27H26FN3O3 | 459.521 | 5 / 2 | 3.8 | Yes |
| 3415 | 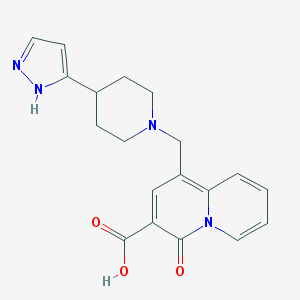 CHEMBL1683363 CHEMBL1683363 | C19H20N4O3 | 352.394 | 5 / 2 | -0.7 | Yes |
| 3423 | 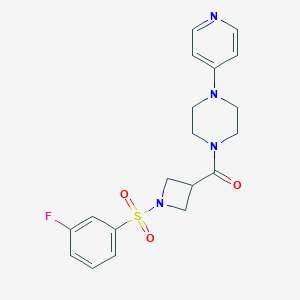 CHEMBL2057492 CHEMBL2057492 | C19H21FN4O3S | 404.46 | 7 / 0 | 0.9 | Yes |
| 3470 |  CHEMBL2333480 CHEMBL2333480 | C27H28F2N4O | 462.545 | 5 / 0 | 3.9 | Yes |
| 3551 |  CHEMBL2312348 CHEMBL2312348 | C22H25NO3 | 351.446 | 4 / 0 | 3.1 | Yes |
| 3639 |  CHEMBL347132 CHEMBL347132 | C25H32N2O2 | 392.543 | 4 / 1 | 3.8 | Yes |
| 3655 |  CHEMBL23620 CHEMBL23620 | C23H23N3O3 | 389.455 | 4 / 1 | 1.7 | Yes |
| 4009 | 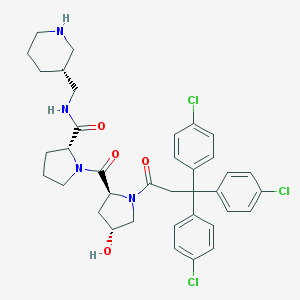 CHEMBL215894 CHEMBL215894 | C37H41Cl3N4O4 | 712.109 | 5 / 3 | 5.9 | No |
| 4321 | 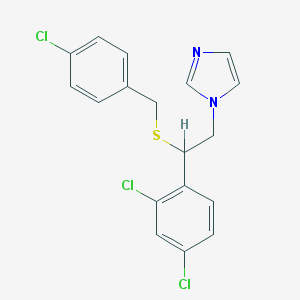 sulconazole sulconazole | C18H15Cl3N2S | 397.742 | 2 / 0 | 6.1 | No |
| 4414 | 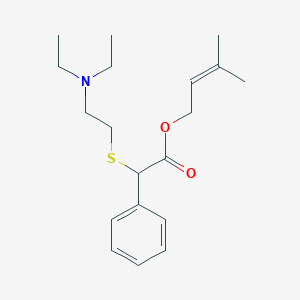 CHEMBL2377264 CHEMBL2377264 | C19H29NO2S | 335.506 | 4 / 0 | 4.8 | Yes |
| 4969 | 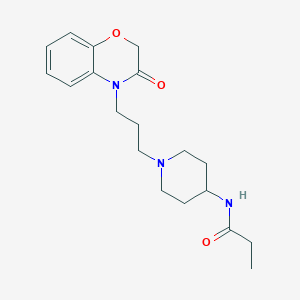 CHEMBL1243232 CHEMBL1243232 | C19H27N3O3 | 345.443 | 4 / 1 | 1.8 | Yes |
| 4985 |  CHEMBL255692 CHEMBL255692 | C36H46Cl2N2O2 | 609.676 | 4 / 4 | N/A | No |
| 5742 |  AHOUBRCZNHFOSL-UHFFFAOYSA-N AHOUBRCZNHFOSL-UHFFFAOYSA-N | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5757 |  paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5893 |  CHEMBL73661 CHEMBL73661 | C28H40N2O2S | 468.7 | 4 / 0 | 6.2 | No |
| 6131 | 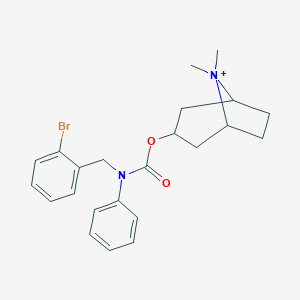 CHEMBL237313 CHEMBL237313 | C23H28BrN2O2+ | 444.393 | 2 / 0 | 5.1 | No |
| 6192 | 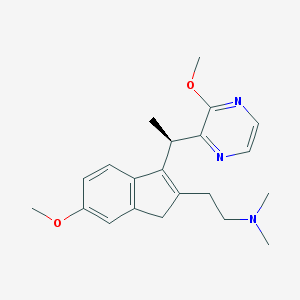 CHEMBL1090186 CHEMBL1090186 | C21H27N3O2 | 353.466 | 5 / 0 | 2.5 | Yes |
| 6249 | 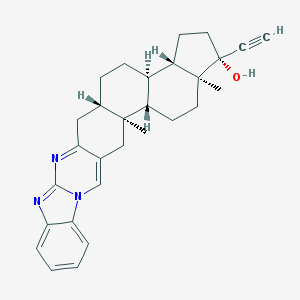 WIN-51708 WIN-51708 | C29H33N3O | 439.603 | 3 / 1 | 6.5 | No |
| 6482 | 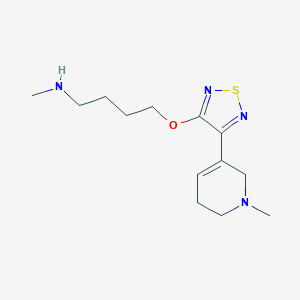 CHEMBL272359 CHEMBL272359 | C13H22N4OS | 282.406 | 6 / 1 | 1.2 | Yes |
| 6610 |  carbachol carbachol | C6H15ClN2O2 | 182.648 | 3 / 1 | N/A | N/A |
| 6681 |  CHEMBL214029 CHEMBL214029 | C38H43F3N4O4 | 676.781 | 8 / 3 | 4.7 | No |
| 6831 |  CHEMBL1940120 CHEMBL1940120 | C19H22N6O3S2 | 446.544 | 9 / 1 | 1.1 | Yes |
| 6833 |  CID 49797317 CID 49797317 | C14H18N2O6 | 310.306 | 8 / 2 | N/A | N/A |
| 6883 |  CHEMBL400038 CHEMBL400038 | C22H26N3O2S+ | 396.529 | 4 / 0 | 3.8 | Yes |
| 463735 |  CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 7185 |  CHEMBL2024329 CHEMBL2024329 | C23H33N3O3 | 399.535 | 4 / 1 | 3.6 | Yes |
| 7188 |  CHEMBL2024331 CHEMBL2024331 | C23H33N3O3 | 399.535 | 4 / 1 | 3.6 | Yes |
| 7296 | 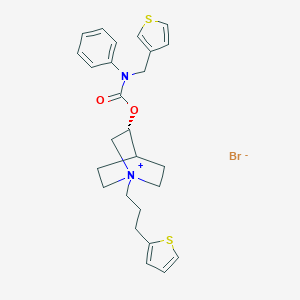 CHEMBL1779132 CHEMBL1779132 | C26H31BrN2O2S2 | 547.57 | 5 / 0 | N/A | No |
| 7414 | 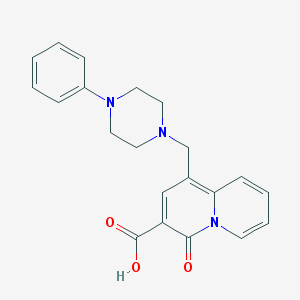 CHEMBL2042688 CHEMBL2042688 | C21H21N3O3 | 363.417 | 5 / 1 | 0.3 | Yes |
| 7487 | 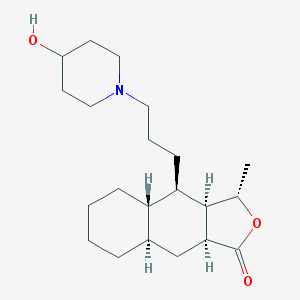 CHEMBL445134 CHEMBL445134 | C21H35NO3 | 349.515 | 4 / 1 | 4.0 | Yes |
| 7654 | 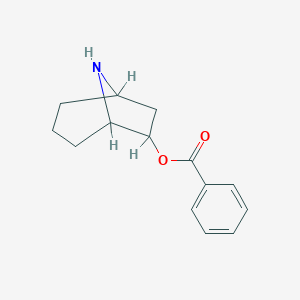 CHEMBL84113 CHEMBL84113 | C14H17NO2 | 231.295 | 3 / 1 | 2.5 | Yes |
| 7779 | 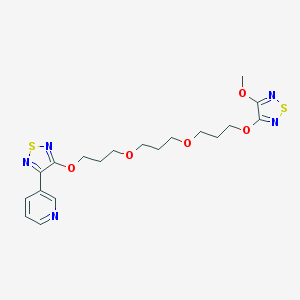 CHEMBL376295 CHEMBL376295 | C19H25N5O5S2 | 467.559 | 12 / 0 | 2.8 | No |
| 7923 | 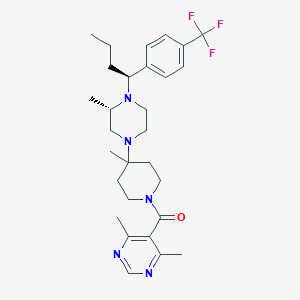 CHEMBL432170 CHEMBL432170 | C29H40F3N5O | 531.668 | 8 / 0 | 5.1 | No |
| 7977 | 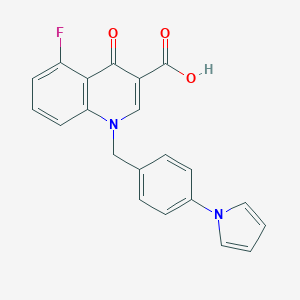 CHEMBL600419 CHEMBL600419 | C21H15FN2O3 | 362.36 | 5 / 1 | 4.0 | Yes |
| 8573 | 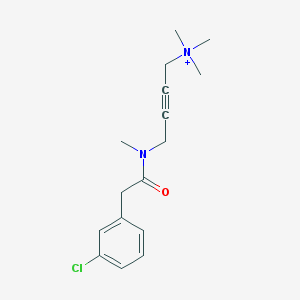 CHEMBL97235 CHEMBL97235 | C16H22ClN2O+ | 293.815 | 1 / 0 | 2.1 | Yes |
| 8674 |  CHEMBL1084807 CHEMBL1084807 | C19H23ClN2O2S | 378.915 | 4 / 1 | 3.6 | Yes |
| 8816 |  CHEMBL2164906 CHEMBL2164906 | C20H24N2O | 308.425 | 2 / 0 | 3.1 | Yes |
| 8817 |  CHEMBL2164911 CHEMBL2164911 | C20H24N2O | 308.425 | 2 / 0 | 3.1 | Yes |
| 8926 |  CHEMBL513228 CHEMBL513228 | C25H32N4O5 | 468.554 | 6 / 4 | 2.7 | Yes |
| 9107 |  CHEMBL307554 CHEMBL307554 | C21H22F2N2O2 | 372.416 | 5 / 3 | 2.7 | Yes |
| 9202 |  CHEMBL519893 CHEMBL519893 | C30H32N6O4 | 540.624 | 6 / 5 | 2.6 | No |
| 9331 |  CHEMBL521244 CHEMBL521244 | C31H33N5O5 | 555.635 | 7 / 4 | 3.6 | No |
| 9420 |  CHEMBL170205 CHEMBL170205 | C21H35NO2 | 333.516 | 3 / 0 | 5.0 | Yes |
| 547991 | 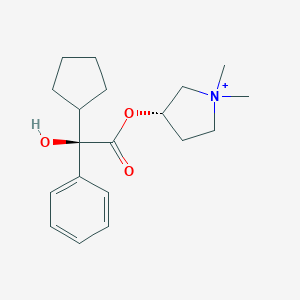 Glycopyrrolate cation Glycopyrrolate cation | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9534 | 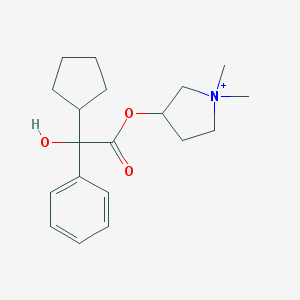 Glycopyrronium Glycopyrronium | C19H28NO3+ | 318.437 | 3 / 1 | 3.1 | Yes |
| 9650 | 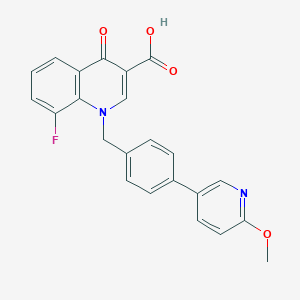 CHEMBL596211 CHEMBL596211 | C23H17FN2O4 | 404.397 | 7 / 1 | 4.4 | Yes |
| 9720 | 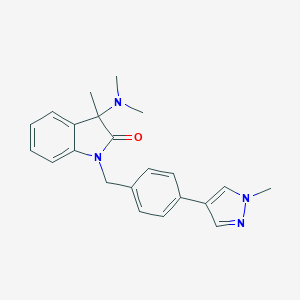 CHEMBL2313495 CHEMBL2313495 | C22H24N4O | 360.461 | 3 / 0 | 2.7 | Yes |
| 442089 |  CHEMBL3414838 CHEMBL3414838 | C30H34BrFN4O4 | 613.528 | 8 / 1 | N/A | No |
| 9857 |  AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 9899 |  CHEMBL168991 CHEMBL168991 | C11H16N2O2 | 208.261 | 4 / 0 | 0.5 | Yes |
| 10003 |  CHEMBL358289 CHEMBL358289 | C26H38F2N2O2 | 448.599 | 5 / 2 | 5.7 | No |
| 10177 | 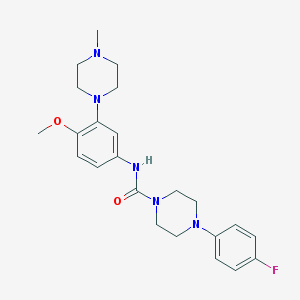 CHEMBL194206 CHEMBL194206 | C23H30FN5O2 | 427.524 | 6 / 1 | 2.7 | Yes |
| 10286 | 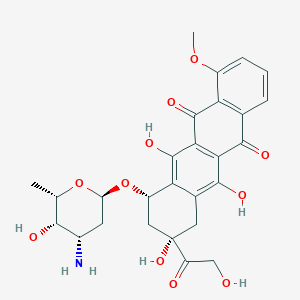 doxorubicin doxorubicin | C27H29NO11 | 543.525 | 12 / 6 | 1.3 | No |
| 10388 | 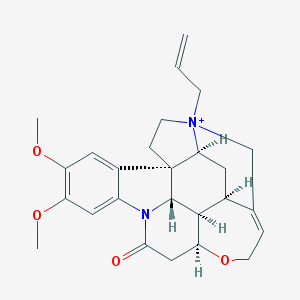 N-Allyl Brucine Iodide N-Allyl Brucine Iodide | C26H31N2O4+ | 435.544 | 4 / 0 | 1.6 | Yes |
| 10462 | 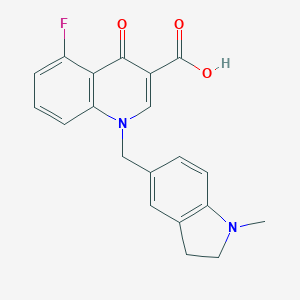 CHEMBL1771699 CHEMBL1771699 | C20H17FN2O3 | 352.365 | 6 / 1 | 3.7 | Yes |
| 10623 | 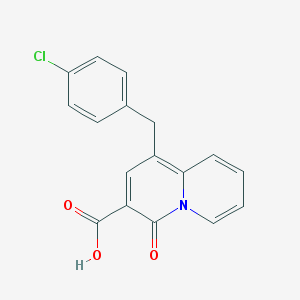 SCHEMBL2145822 SCHEMBL2145822 | C17H12ClNO3 | 313.737 | 3 / 1 | 3.7 | Yes |
| 10707 | 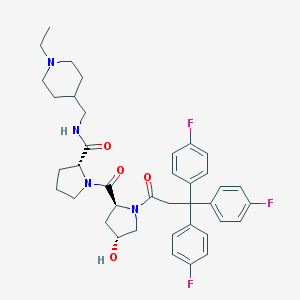 CHEMBL213709 CHEMBL213709 | C39H45F3N4O4 | 690.808 | 8 / 2 | 5.1 | No |
| 10720 | 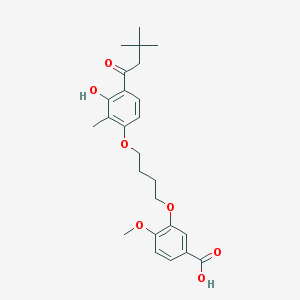 CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 10827 | 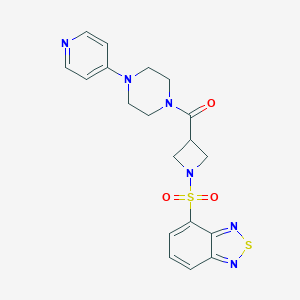 CHEMBL2058897 CHEMBL2058897 | C19H20N6O3S2 | 444.528 | 9 / 0 | 0.7 | Yes |
| 10913 | 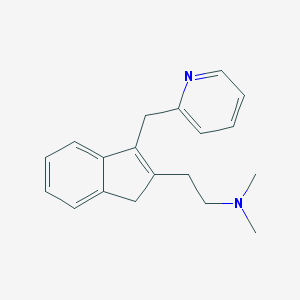 CHEMBL169494 CHEMBL169494 | C19H22N2 | 278.399 | 2 / 0 | 2.4 | Yes |
| 11008 | 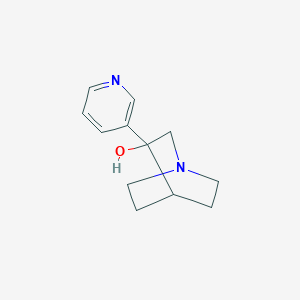 CHEMBL423746 CHEMBL423746 | C12H16N2O | 204.273 | 3 / 1 | 0.4 | Yes |
| 11182 | 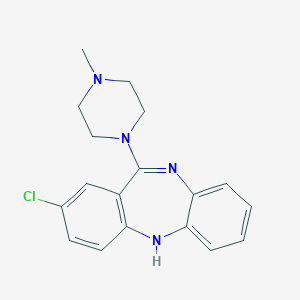 ISOCLOZAPINE ISOCLOZAPINE | C18H19ClN4 | 326.828 | 3 / 1 | 3.1 | Yes |
| 11384 | 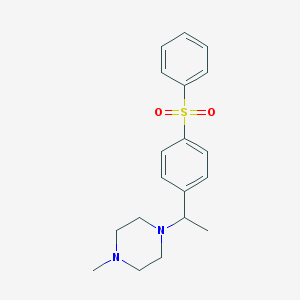 CHEMBL73410 CHEMBL73410 | C19H24N2O2S | 344.473 | 4 / 0 | 2.8 | Yes |
| 521788 | 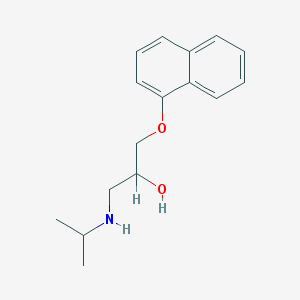 propranolol propranolol | C16H21NO2 | 259.349 | 3 / 2 | 3.0 | Yes |
| 11763 | 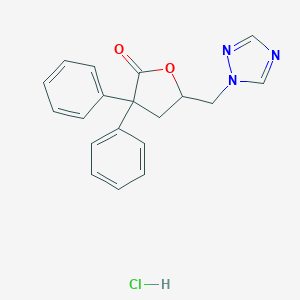 CHEMBL556590 CHEMBL556590 | C19H18ClN3O2 | 355.822 | 4 / 1 | N/A | N/A |
| 11781 | 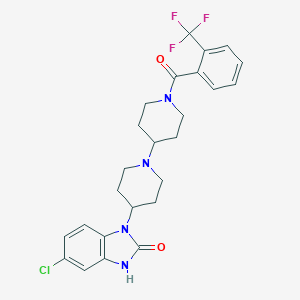 CHEMBL521388 CHEMBL521388 | C25H26ClF3N4O2 | 506.954 | 6 / 1 | 4.6 | No |
| 11915 | 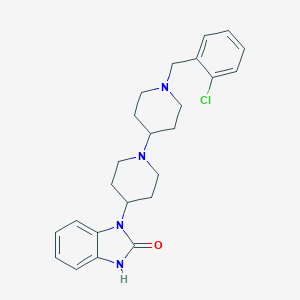 CHEMBL477821 CHEMBL477821 | C24H29ClN4O | 424.973 | 3 / 1 | 4.1 | Yes |
| 11990 | 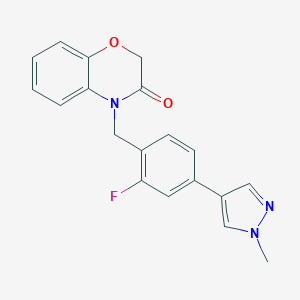 CHEMBL2313512 CHEMBL2313512 | C19H16FN3O2 | 337.354 | 4 / 0 | 2.6 | Yes |
| 12118 | 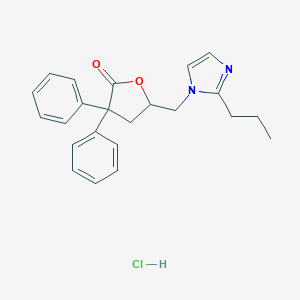 CHEMBL555173 CHEMBL555173 | C23H25ClN2O2 | 396.915 | 3 / 1 | N/A | N/A |
| 464367 | 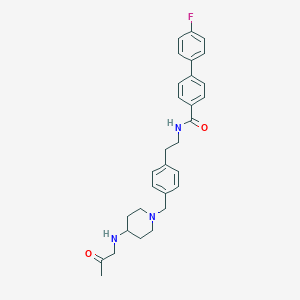 CHEMBL3600817 CHEMBL3600817 | C30H34FN3O2 | 487.619 | 5 / 2 | 4.6 | Yes |
| 12422 | 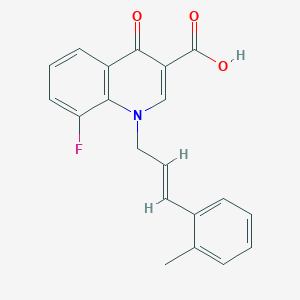 CHEMBL1771772 CHEMBL1771772 | C20H16FNO3 | 337.35 | 5 / 1 | 4.6 | Yes |
| 12543 | 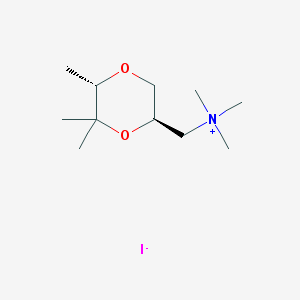 CHEMBL593620 CHEMBL593620 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12548 | 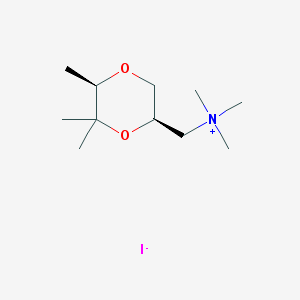 CHEMBL593685 CHEMBL593685 | C11H24INO2 | 329.222 | 3 / 0 | N/A | N/A |
| 12597 | 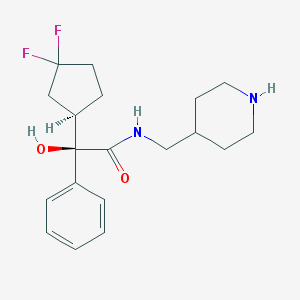 CHEMBL306903 CHEMBL306903 | C19H26F2N2O2 | 352.426 | 5 / 3 | 2.5 | Yes |
| 12674 | 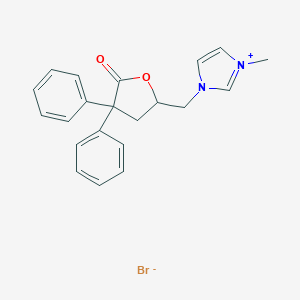 CHEMBL343112 CHEMBL343112 | C21H21BrN2O2 | 413.315 | 3 / 0 | N/A | N/A |
| 12701 | 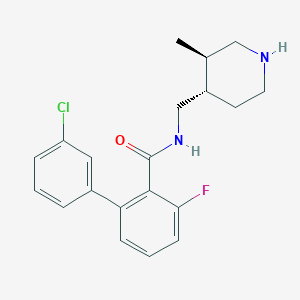 CHEMBL1083925 CHEMBL1083925 | C20H22ClFN2O | 360.857 | 3 / 2 | 4.3 | Yes |
| 12705 | 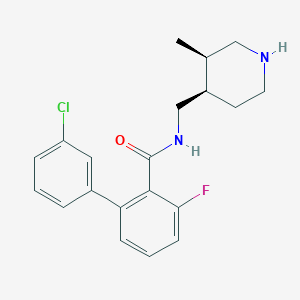 CHEMBL1083926 CHEMBL1083926 | C20H22ClFN2O | 360.857 | 3 / 2 | 4.3 | Yes |
| 12725 | 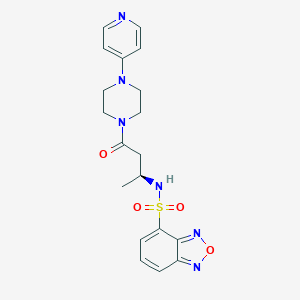 CHEMBL1940135 CHEMBL1940135 | C19H22N6O4S | 430.483 | 9 / 1 | 0.5 | Yes |
| 12751 | 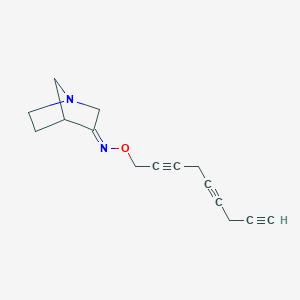 CHEMBL83074 CHEMBL83074 | C15H16N2O | 240.306 | 3 / 0 | 2.1 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417