

You can:
| Name | Muscarinic acetylcholine receptor DM1 |
|---|---|
| Species | Drosophila melanogaster (Fruit fly) |
| Gene | mAChR-A |
| Synonym | N/A |
| Disease | N/A for non-human GPCRs |
| Length | 805 |
| Amino acid sequence | MEPVMSLALAAHGPPSILEPLFKTVTTSTTTTTTTTTSTTTTTASPAGYSPGYPGTTLLTALFENLTSTAASGLYDPYSGMYGNQTNGTIGFETKGPRYSLASMVVMGFVAAILSTVTVAGNVMVMISFKIDKQLQTISNYFLFSLAIADFAIGAISMPLFAVTTILGYWPLGPIVCDTWLALDYLASNASVLNLLIISFDRYFSVTRPLTYRAKRTTNRAAVMIGAAWGISLLLWPPWIYSWPYIEGKRTVPKDECYIQFIETNQYITFGTALAAFYFPVTIMCFLYWRIWRETKKRQKDLPNLQAGKKDSSKRSNSSDENTVVNHASGGLLAFAQVGGNDHDTWRRPRSESSPDAESVYMTNMVIDSGYHGMHSRKSSIKSTNTIKKSYTCFGSIKEWCIAWWHSGREDSDDFAYEQEEPSDLGYATPVTIETPLQSSVSRCTSMNVMRDNYSMGGSVSGVRPPSILLSDVSPTPLPRPPLASISQLQEMSAVTASTTANVNTSGNGNGAINNNNNASHNGNGAVNGNGAGNGSGIGLGTTGNATHRDSRTLPVINRINSRSVSQDSVYTILIRLPSDGASSNAANGGGGGPGAGAAASASLSMQGDCAPSIKMIHEDGPTTTAAAAPLASAAATRRPLPSRDSEFSLPLGRRMSHAQHDARLLNAKVIPKQLGKAGGGAAGGGVGGAHALMNARNAAKKKKKSQEKRQESKAAKTLSAILLSFIITWTPYNILVLIKPLTTCSDCIPTELWDFFYALCYINSTINPMCYALCNATFRRTYVRILTCKWHTRNREGMVRGVYN |
| UniProt | P16395 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | N/A |
| 3D structure model | No available structures or models |
| BioLiP | N/A |
| Therapeutic Target Database | N/A |
| ChEMBL | CHEMBL2366467 |
| IUPHAR | N/A |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 39354 | 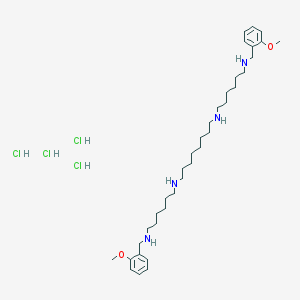 Methoctramine Methoctramine | C36H66Cl4N4O2 | 728.75 | 6 / 8 | N/A | No |
| 43846 | 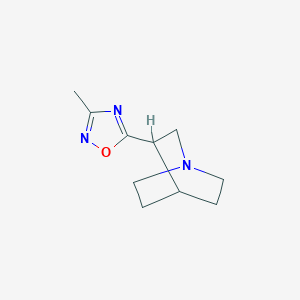 114724-64-0 114724-64-0 | C10H15N3O | 193.25 | 4 / 0 | 1.2 | Yes |
| 45429 | 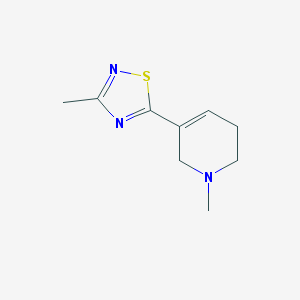 CHEMBL304873 CHEMBL304873 | C9H13N3S | 195.284 | 4 / 0 | 1.4 | Yes |
| 114238 | 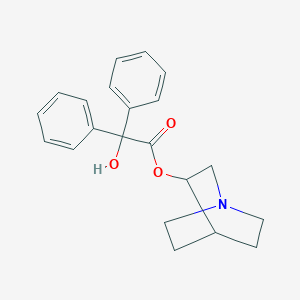 3-Quinuclidinyl benzilate 3-Quinuclidinyl benzilate | C21H23NO3 | 337.419 | 4 / 1 | 3.3 | Yes |
| 133542 | 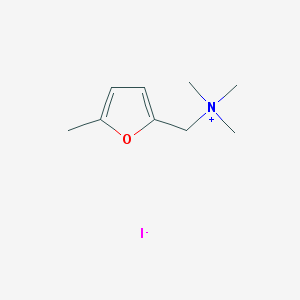 5-METHYLFURMETHIODIDE 5-METHYLFURMETHIODIDE | C9H16INO | 281.137 | 2 / 0 | N/A | N/A |
| 183188 | 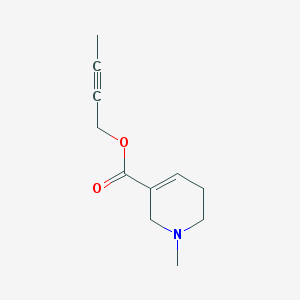 AC1N6FGH AC1N6FGH | C11H15NO2 | 193.246 | 3 / 0 | 1.0 | Yes |
| 215165 | 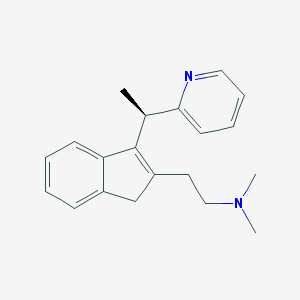 R-dimethindene R-dimethindene | C20H24N2 | 292.426 | 2 / 0 | 2.7 | Yes |
| 217762 | 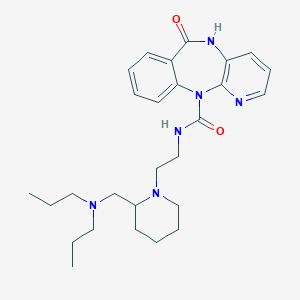 AF-DX 384 AF-DX 384 | C27H38N6O2 | 478.641 | 5 / 2 | 2.8 | Yes |
| 250013 | 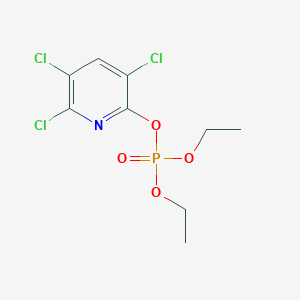 Chlorpyrifos oxon Chlorpyrifos oxon | C9H11Cl3NO4P | 334.514 | 5 / 0 | 3.5 | Yes |
| 262836 | 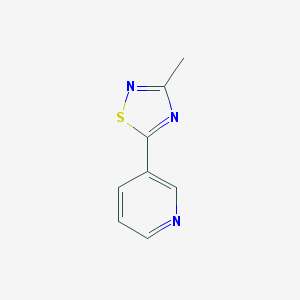 92751-39-8 92751-39-8 | C8H7N3S | 177.225 | 4 / 0 | 1.7 | Yes |
| 274812 | 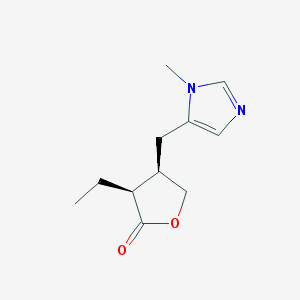 pilocarpine pilocarpine | C11H16N2O2 | 208.261 | 3 / 0 | 1.1 | Yes |
| 299735 | 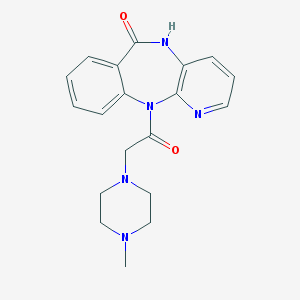 pirenzepine pirenzepine | C19H21N5O2 | 351.41 | 5 / 1 | 0.1 | Yes |
| 303766 | 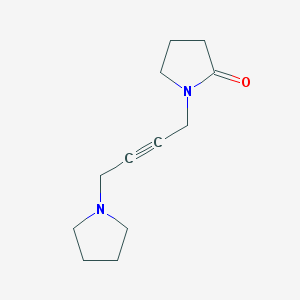 Oxotremorine Oxotremorine | C12H18N2O | 206.289 | 2 / 0 | 0.4 | Yes |
| 319939 | 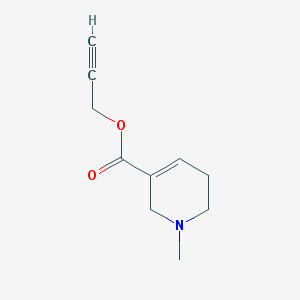 Arecaidine propargyl ester Arecaidine propargyl ester | C10H13NO2 | 179.219 | 3 / 0 | 0.5 | Yes |
| 332909 | 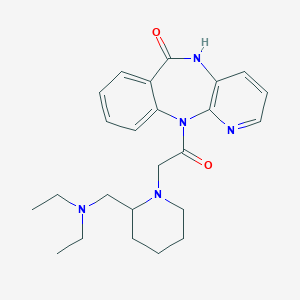 Otenzepad Otenzepad | C24H31N5O2 | 421.545 | 5 / 1 | 2.0 | Yes |
| 352163 | 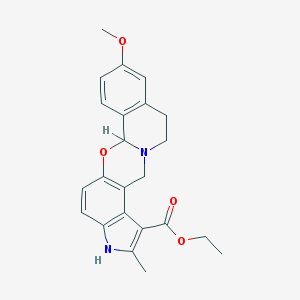 PD 102807 PD 102807 | C23H24N2O4 | 392.455 | 5 / 1 | 3.9 | Yes |
| 380470 | 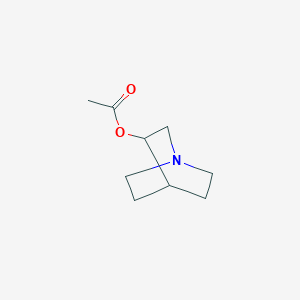 aceclidine aceclidine | C9H15NO2 | 169.224 | 3 / 0 | 0.7 | Yes |
| 383953 | 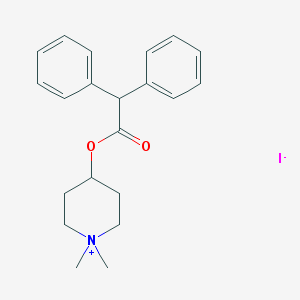 4-Damp methiodide 4-Damp methiodide | C21H26INO2 | 451.348 | 3 / 0 | N/A | N/A |
| 385507 | 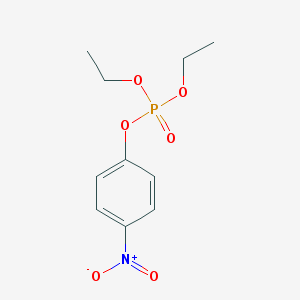 PARAOXON PARAOXON | C10H14NO6P | 275.197 | 6 / 0 | 2.0 | Yes |
| 421163 | 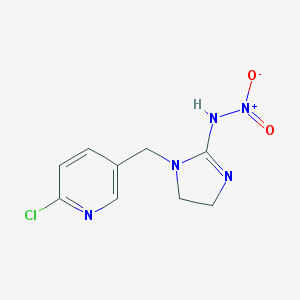 1-((6-Chloro-3-pyridinyl)methyl)-4,5-dihydro-N-nitro-imidazol-2-amine 1-((6-Chloro-3-pyridinyl)methyl)-4,5-dihydro-N-nitro-imidazol-2-amine | C9H10ClN5O2 | 255.662 | 4 / 1 | 0.8 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417