

You can:
| Name | Substance-K receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | TACR2 |
| Synonym | NK2 receptor Neurokinin A receptor Tachykinin receptor 2 TAC2R Substance K receptor [ Show all ] |
| Disease | Urinary incontinence Pain Unspecified Irritable bowel syndrome with diarrhoea Irritable bowel syndrome [ Show all ] |
| Length | 398 |
| Amino acid sequence | MGTCDIVTEANISSGPESNTTGITAFSMPSWQLALWATAYLALVLVAVTGNAIVIWIILAHRRMRTVTNYFIVNLALADLCMAAFNAAFNFVYASHNIWYFGRAFCYFQNLFPITAMFVSIYSMTAIAADRYMAIVHPFQPRLSAPSTKAVIAGIWLVALALASPQCFYSTVTMDQGATKCVVAWPEDSGGKTLLLYHLVVIALIYFLPLAVMFVAYSVIGLTLWRRAVPGHQAHGANLRHLQAMKKFVKTMVLVVLTFAICWLPYHLYFILGSFQEDIYCHKFIQQVYLALFWLAMSSTMYNPIIYCCLNHRFRSGFRLAFRCCPWVTPTKEDKLELTPTTSLSTRVNRCHTKETLFMAGDTAPSEATSGEAGRPQDGSGLWFGYGLLAPTKTHVEI |
| UniProt | P21452 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P21452 |
| 3D structure model | This predicted structure model is from GPCR-EXP P21452. |
| BioLiP | N/A |
| Therapeutic Target Database | T52790 |
| ChEMBL | CHEMBL2327 |
| IUPHAR | 361 |
| DrugBank | BE0002222 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 760 | 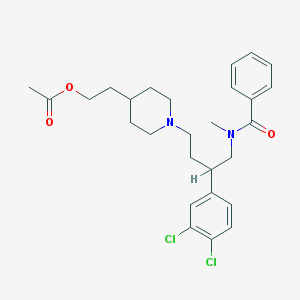 CHEMBL57589 CHEMBL57589 | C27H34Cl2N2O3 | 505.48 | 4 / 0 | 5.8 | No |
| 890 | 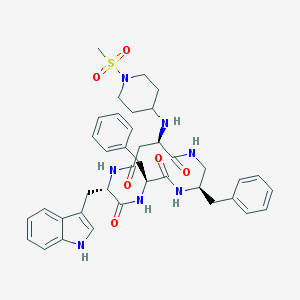 CHEMBL439281 CHEMBL439281 | C39H47N7O6S | 741.908 | 8 / 6 | 2.9 | No |
| 1557 | 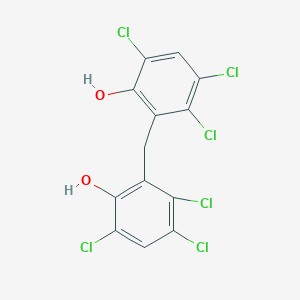 hexachlorophene hexachlorophene | C13H6Cl6O2 | 406.889 | 2 / 2 | 7.5 | No |
| 2064 | 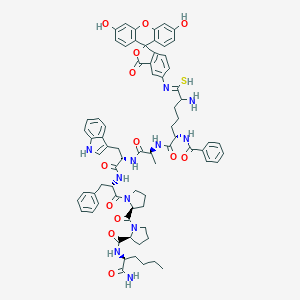 CHEMBL413205 CHEMBL413205 | C73H79N11O13S | 1350.56 | 16 / 11 | 7.6 | No |
| 2252 |  CHEMBL236661 CHEMBL236661 | C31H40N4O3S | 548.746 | 5 / 4 | 5.3 | No |
| 2324 |  CHEMBL244766 CHEMBL244766 | C32H42N4O5S | 594.771 | 7 / 6 | 3.6 | No |
| 2495 |  Substance P Substance P | C63H98N18O13S | 1347.65 | 17 / 15 | -2.3 | No |
| 2800 |  sub-stance p sub-stance p | C63H98N18O13S | 1347.65 | 17 / 15 | -2.3 | No |
| 3199 | 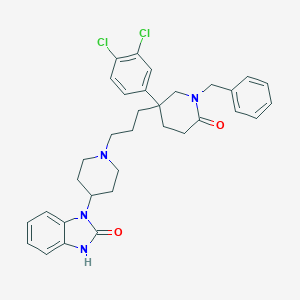 CHEMBL26790 CHEMBL26790 | C33H36Cl2N4O2 | 591.577 | 3 / 1 | 6.1 | No |
| 3516 | 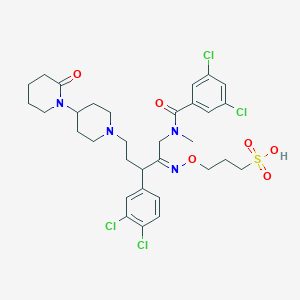 CHEMBL166827 CHEMBL166827 | C32H40Cl4N4O6S | 750.554 | 8 / 1 | 3.7 | No |
| 3694 | 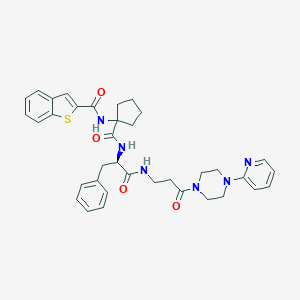 CHEMBL242626 CHEMBL242626 | C36H40N6O4S | 652.814 | 7 / 3 | 4.6 | No |
| 3846 | 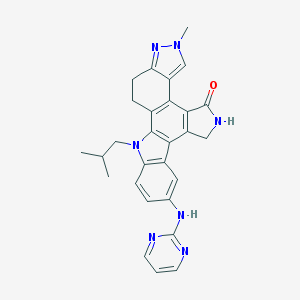 CEP-11981 CEP-11981 | C28H27N7O | 477.572 | 5 / 2 | 3.9 | Yes |
| 4325 | 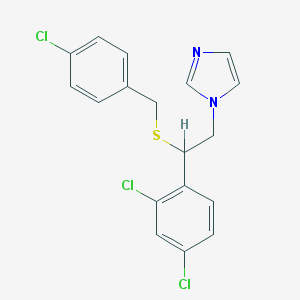 sulconazole sulconazole | C18H15Cl3N2S | 397.742 | 2 / 0 | 6.1 | No |
| 4837 | 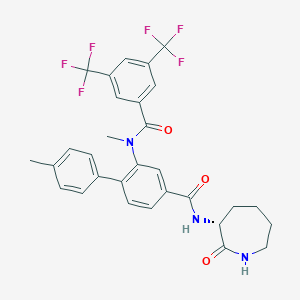 CHEMBL2112211 CHEMBL2112211 | C30H27F6N3O3 | 591.554 | 9 / 2 | 6.2 | No |
| 5406 | 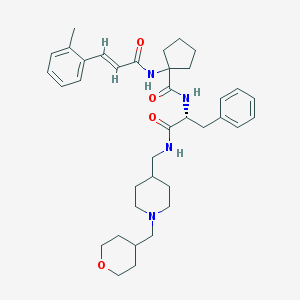 CHEMBL446604 CHEMBL446604 | C37H50N4O4 | 614.831 | 5 / 3 | 5.1 | No |
| 6046 | 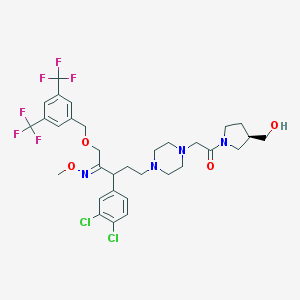 CHEMBL75367 CHEMBL75367 | C32H38Cl2F6N4O4 | 727.57 | 13 / 1 | 6.0 | No |
| 6048 | 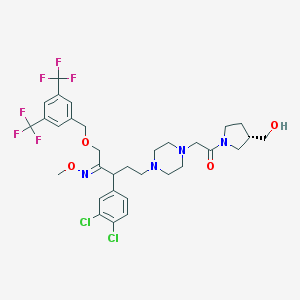 CHEMBL75436 CHEMBL75436 | C32H38Cl2F6N4O4 | 727.57 | 13 / 1 | 6.0 | No |
| 6165 | 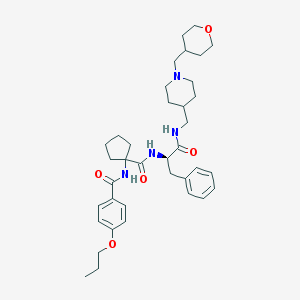 CHEMBL499689 CHEMBL499689 | C37H52N4O5 | 632.846 | 6 / 3 | 5.2 | No |
| 6655 | 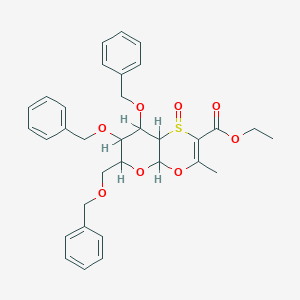 CID 10077178 CID 10077178 | C33H36O8S | 592.703 | 9 / 0 | 4.2 | No |
| 7265 | 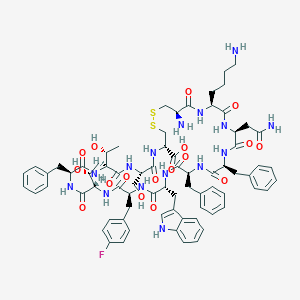 CHEMBL2112245 CHEMBL2112245 | C74H92FN15O17S2 | 1546.76 | 22 / 19 | -0.3 | No |
| 7432 | 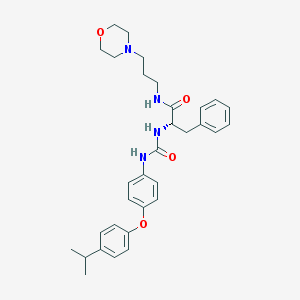 CHEMBL441115 CHEMBL441115 | C32H40N4O4 | 544.696 | 5 / 3 | 4.9 | No |
| 7433 | 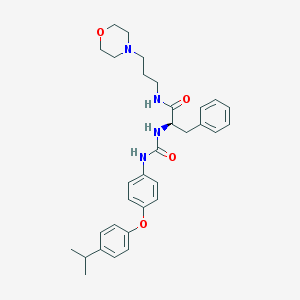 CHEMBL362201 CHEMBL362201 | C32H40N4O4 | 544.696 | 5 / 3 | 4.9 | No |
| 7866 | 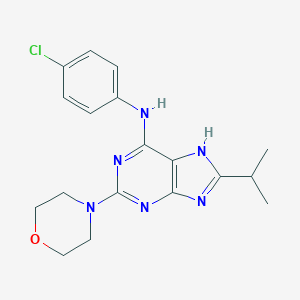 CHEMBL98977 CHEMBL98977 | C18H21ClN6O | 372.857 | 6 / 2 | 3.8 | Yes |
| 7952 | 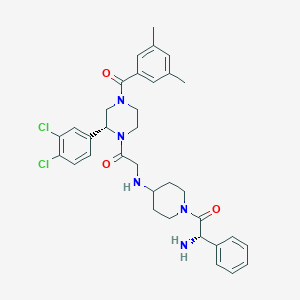 CHEMBL106300 CHEMBL106300 | C34H39Cl2N5O3 | 636.618 | 5 / 2 | 4.6 | No |
| 7955 |  CHEMBL107306 CHEMBL107306 | C34H39Cl2N5O3 | 636.618 | 5 / 2 | 4.6 | No |
| 8034 |  CHEMBL2370513 CHEMBL2370513 | C42H54N8O9 | 814.941 | 9 / 8 | 2.9 | No |
| 8652 |  CHEMBL359885 CHEMBL359885 | C25H34N4O3 | 438.572 | 4 / 2 | 2.4 | Yes |
| 8653 |  CHEMBL181998 CHEMBL181998 | C25H34N4O3 | 438.572 | 4 / 2 | 2.4 | Yes |
| 9141 | 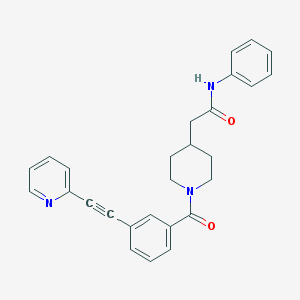 CHEMBL208900 CHEMBL208900 | C27H25N3O2 | 423.516 | 3 / 1 | 3.9 | Yes |
| 9452 | 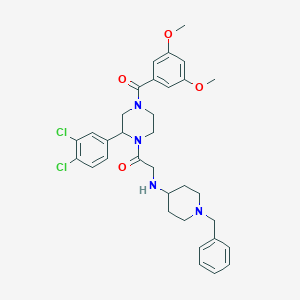 CHEMBL319474 CHEMBL319474 | C33H38Cl2N4O4 | 625.591 | 6 / 1 | 5.1 | No |
| 9873 | 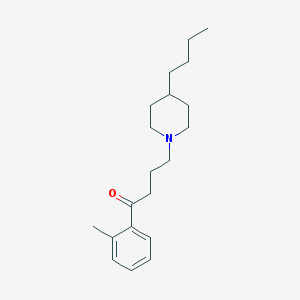 AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 9998 | 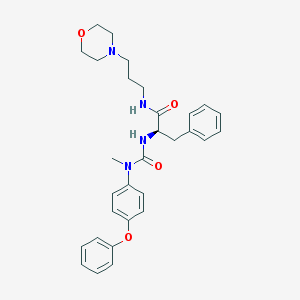 CHEMBL180238 CHEMBL180238 | C30H36N4O4 | 516.642 | 5 / 2 | 4.0 | No |
| 9999 | 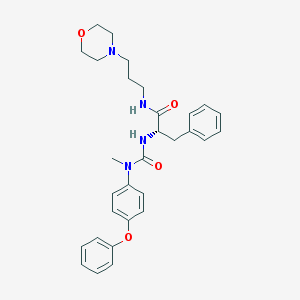 CHEMBL180001 CHEMBL180001 | C30H36N4O4 | 516.642 | 5 / 2 | 4.0 | No |
| 10247 | 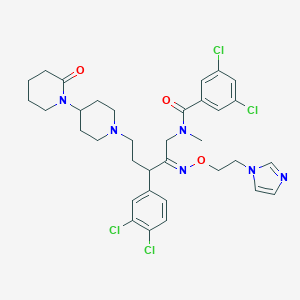 CHEMBL168144 CHEMBL168144 | C34H40Cl4N6O3 | 722.533 | 6 / 0 | 6.4 | No |
| 521745 | 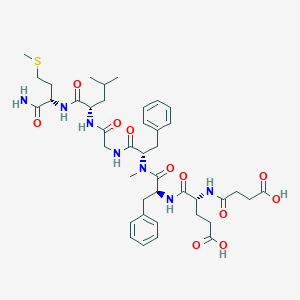 CHEMBL3735396 CHEMBL3735396 | C41H57N7O11S | 856.005 | 12 / 8 | 1.5 | No |
| 521747 | 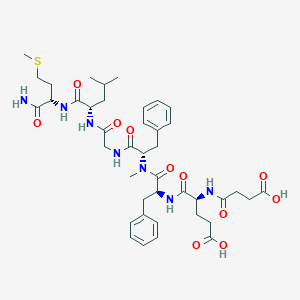 CHEMBL3735210 CHEMBL3735210 | C41H57N7O11S | 856.005 | 12 / 8 | 1.5 | No |
| 10951 | 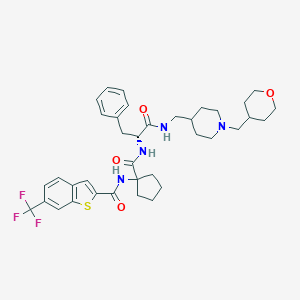 CHEMBL395972 CHEMBL395972 | C37H45F3N4O4S | 698.846 | 9 / 3 | 6.6 | No |
| 11073 | 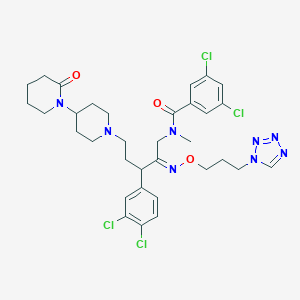 CHEMBL287369 CHEMBL287369 | C33H40Cl4N8O3 | 738.536 | 8 / 0 | 6.4 | No |
| 12446 | 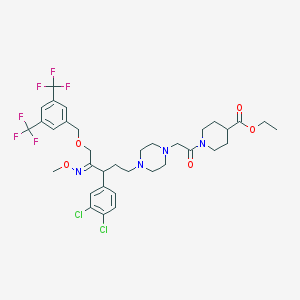 CHEMBL41068 CHEMBL41068 | C35H42Cl2F6N4O5 | 783.634 | 14 / 0 | 7.0 | No |
| 521829 | 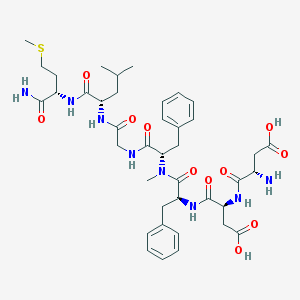 CHEMBL3736313 CHEMBL3736313 | C40H56N8O11S | 856.993 | 13 / 9 | -2.0 | No |
| 12807 | 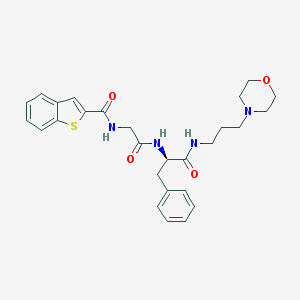 CHEMBL242846 CHEMBL242846 | C27H32N4O4S | 508.637 | 6 / 3 | 3.3 | No |
| 13057 | 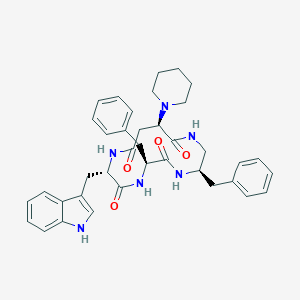 CHEMBL387699 CHEMBL387699 | C38H44N6O4 | 648.808 | 5 / 5 | 4.3 | No |
| 13364 | 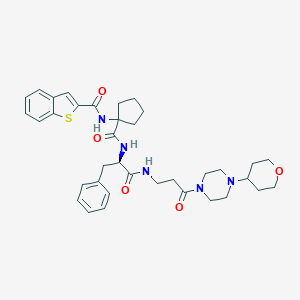 CHEMBL394607 CHEMBL394607 | C36H45N5O5S | 659.846 | 7 / 3 | 3.9 | No |
| 13713 | 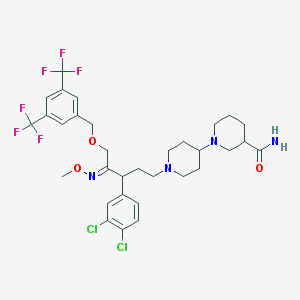 CHEMBL342341 CHEMBL342341 | C32H38Cl2F6N4O3 | 711.571 | 12 / 1 | 6.9 | No |
| 14108 | 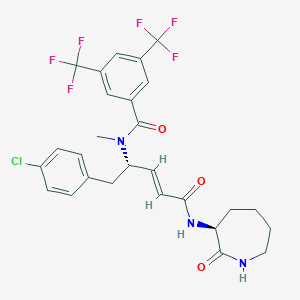 CHEMBL2114398 CHEMBL2114398 | C27H26ClF6N3O3 | 589.963 | 9 / 2 | 5.6 | No |
| 14109 | 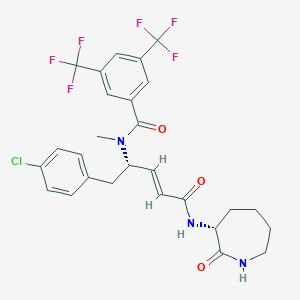 CHEMBL2112070 CHEMBL2112070 | C27H26ClF6N3O3 | 589.963 | 9 / 2 | 5.6 | No |
| 14112 | 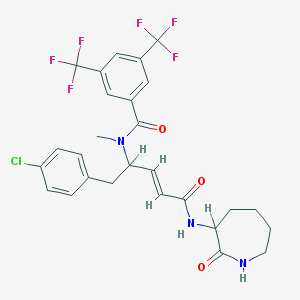 CHEMBL35857 CHEMBL35857 | C27H26ClF6N3O3 | 589.963 | 9 / 2 | 5.6 | No |
| 14114 | 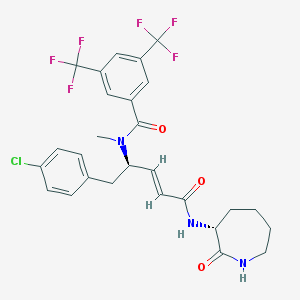 CHEMBL104954 CHEMBL104954 | C27H26ClF6N3O3 | 589.963 | 9 / 2 | 5.6 | No |
| 14116 | 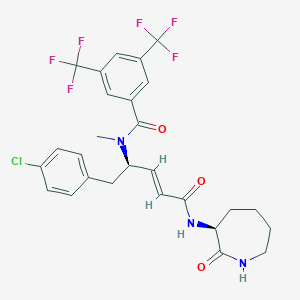 CHEMBL2115343 CHEMBL2115343 | C27H26ClF6N3O3 | 589.963 | 9 / 2 | 5.6 | No |
| 14881 | 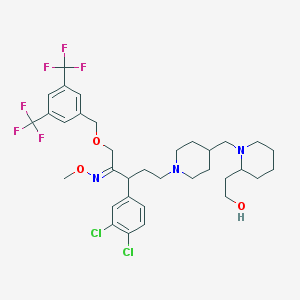 CHEMBL145302 CHEMBL145302 | C34H43Cl2F6N3O3 | 726.626 | 12 / 1 | 8.3 | No |
| 15231 | 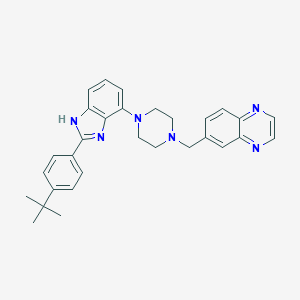 WAY-207024 WAY-207024 | C30H32N6 | 476.628 | 5 / 1 | 5.5 | No |
| 15241 | 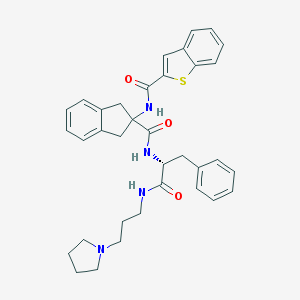 CHEMBL394420 CHEMBL394420 | C35H38N4O3S | 594.774 | 5 / 3 | 6.0 | No |
| 15333 | 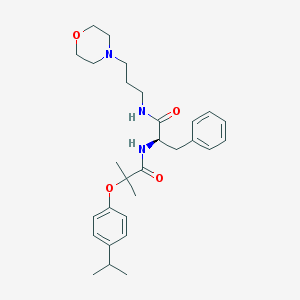 CHEMBL179886 CHEMBL179886 | C29H41N3O4 | 495.664 | 5 / 2 | 4.3 | Yes |
| 15334 | 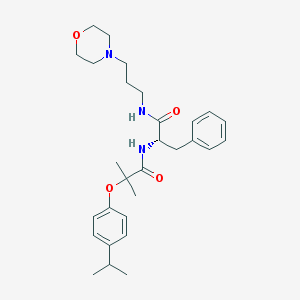 CHEMBL179891 CHEMBL179891 | C29H41N3O4 | 495.664 | 5 / 2 | 4.3 | Yes |
| 521933 | 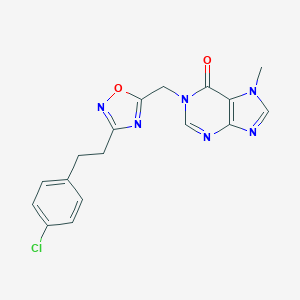 AM-0902 AM-0902 | C17H15ClN6O2 | 370.797 | 6 / 0 | 2.3 | Yes |
| 548062 | 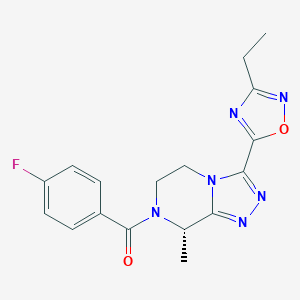 CHEMBL3984151 CHEMBL3984151 | C17H17FN6O2 | 356.361 | 7 / 0 | 1.4 | Yes |
| 557729 | 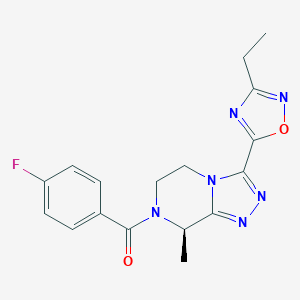 SCHEMBL16143091 SCHEMBL16143091 | C17H17FN6O2 | 356.361 | 7 / 0 | 1.4 | Yes |
| 536393 | 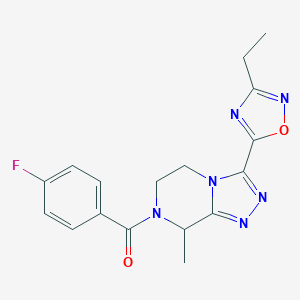 CHEMBL3942295 CHEMBL3942295 | C17H17FN6O2 | 356.361 | 7 / 0 | 1.4 | Yes |
| 16172 | 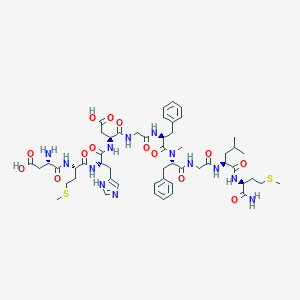 CHEMBL2347498 CHEMBL2347498 | C53H75N13O14S2 | 1182.38 | 18 / 13 | -2.7 | No |
| 16538 | 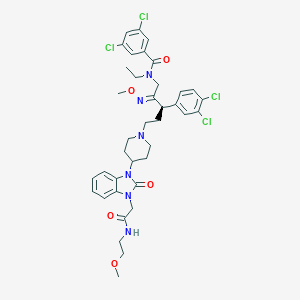 CHEMBL75478 CHEMBL75478 | C38H44Cl4N6O5 | 806.607 | 7 / 1 | 7.1 | No |
| 16665 | 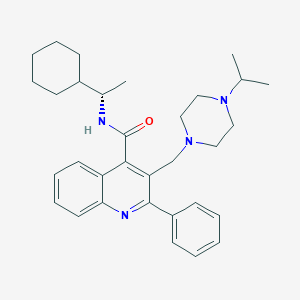 CHEMBL47775 CHEMBL47775 | C32H42N4O | 498.715 | 4 / 1 | 6.4 | No |
| 16902 | 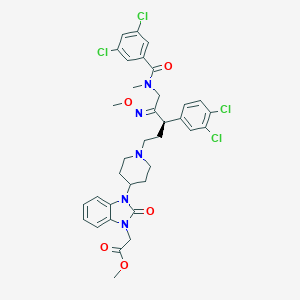 CHEMBL78851 CHEMBL78851 | C35H37Cl4N5O5 | 749.511 | 7 / 0 | 7.4 | No |
| 16948 | 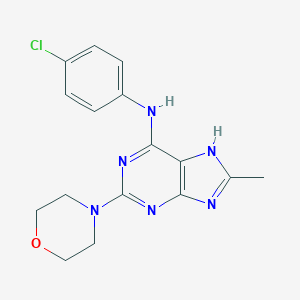 CHEMBL327531 CHEMBL327531 | C16H17ClN6O | 344.803 | 6 / 2 | 3.1 | Yes |
| 17231 | 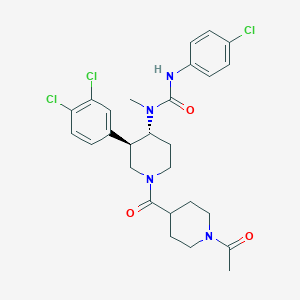 SCHEMBL1973042 SCHEMBL1973042 | C27H31Cl3N4O3 | 565.92 | 3 / 1 | 4.4 | No |
| 17565 | 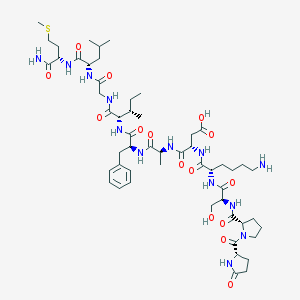 ELEDOISIN ELEDOISIN | C54H85N13O15S | 1188.41 | 17 / 14 | -3.6 | No |
| 17863 | 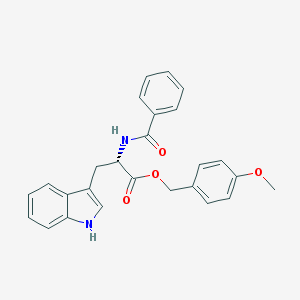 CHEMBL282728 CHEMBL282728 | C26H24N2O4 | 428.488 | 4 / 2 | 4.5 | Yes |
| 18002 | 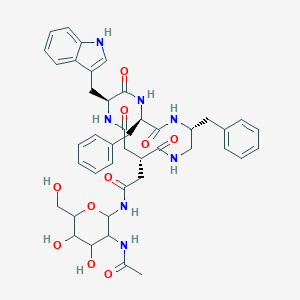 CHEMBL161288 CHEMBL161288 | C43H51N7O10 | 825.92 | 10 / 10 | 0.5 | No |
| 18004 | 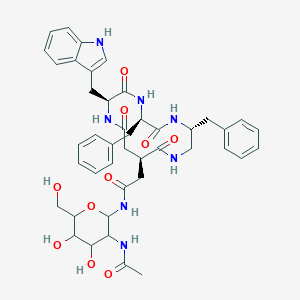 CHEMBL350596 CHEMBL350596 | C43H51N7O10 | 825.92 | 10 / 10 | 0.5 | No |
| 18059 | 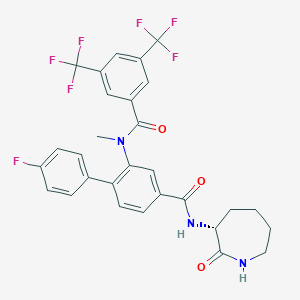 CHEMBL2113739 CHEMBL2113739 | C29H24F7N3O3 | 595.518 | 10 / 2 | 5.9 | No |
| 18973 | 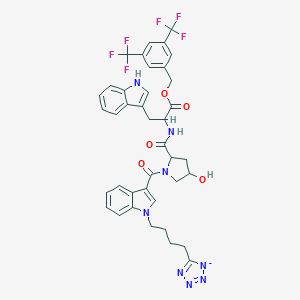 CHEMBL65468 CHEMBL65468 | C39H35F6N8O5- | 809.75 | 15 / 3 | 5.2 | No |
| 19022 | 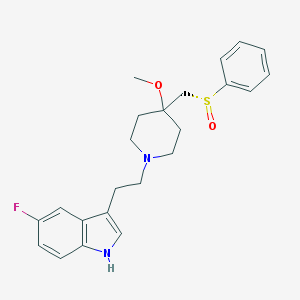 GR-159897 GR-159897 | C23H27FN2O2S | 414.539 | 5 / 1 | 3.4 | Yes |
| 19025 | 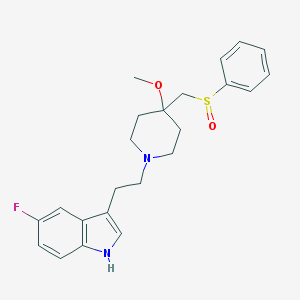 CHEMBL166161 CHEMBL166161 | C23H27FN2O2S | 414.539 | 5 / 1 | 3.4 | Yes |
| 20032 | 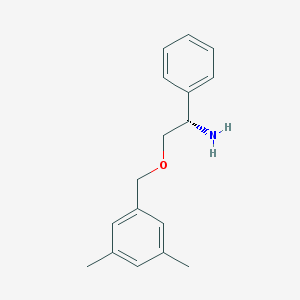 CHEMBL71498 CHEMBL71498 | C17H21NO | 255.361 | 2 / 1 | 2.9 | Yes |
| 20320 | 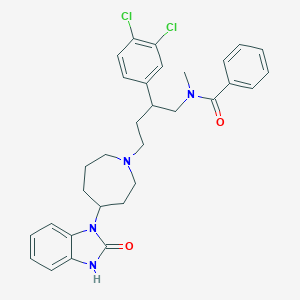 CHEMBL74127 CHEMBL74127 | C31H34Cl2N4O2 | 565.539 | 3 / 1 | 6.2 | No |
| 20724 | 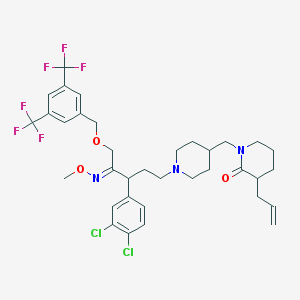 CHEMBL144011 CHEMBL144011 | C35H41Cl2F6N3O3 | 736.621 | 11 / 0 | 8.8 | No |
| 21021 | 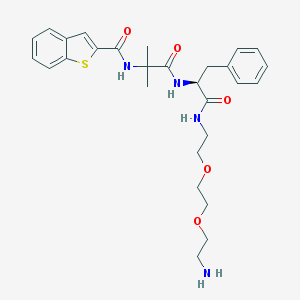 CHEMBL236659 CHEMBL236659 | C28H36N4O5S | 540.679 | 7 / 4 | 2.6 | No |
| 21500 | 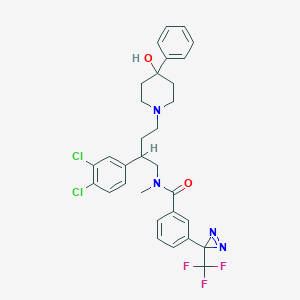 CHEMBL348599 CHEMBL348599 | C31H31Cl2F3N4O2 | 619.51 | 8 / 1 | 6.9 | No |
| 21726 | 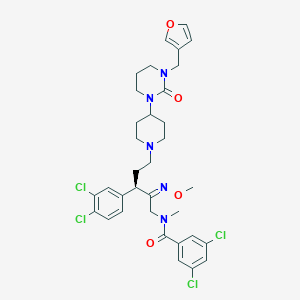 CHEMBL307733 CHEMBL307733 | C34H39Cl4N5O4 | 723.517 | 6 / 0 | 7.0 | No |
| 21976 | 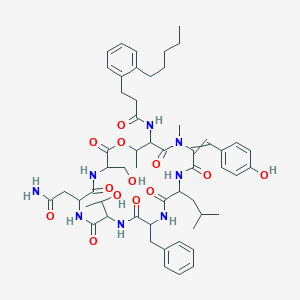 L-Serine,N-[1-oxo-3-(2-pentylphenyl)propyl]-L-threonyl-(aE)-a,b-didehydro-N-methyltyrosyl-L-leucyl-D-phenylalanyl-L-allothreonyl-L-asparaginyl-,(7®1)-lactone L-Serine,N-[1-oxo-3-(2-pentylphenyl)propyl]-L-threonyl-(aE)-a,b-didehydro-N-methyltyrosyl-L-leucyl-D-phenylalanyl-L-allothreonyl-L-asparaginyl-,(7®1)-lactone | C54H72N8O13 | 1041.21 | 13 / 10 | 4.9 | No |
| 522129 | 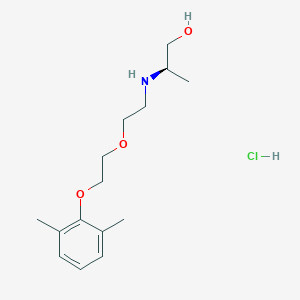 CHEMBL3775436 CHEMBL3775436 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 522177 | 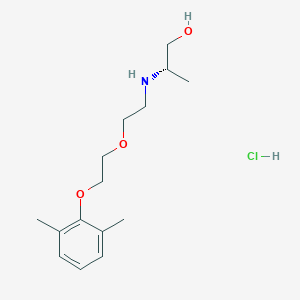 CHEMBL3775769 CHEMBL3775769 | C15H26ClNO3 | 303.827 | 4 / 3 | N/A | N/A |
| 22183 | 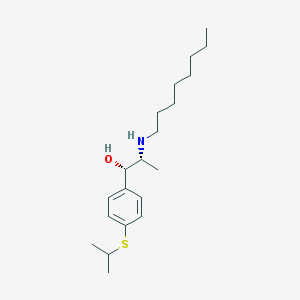 suloctidil suloctidil | C20H35NOS | 337.566 | 3 / 2 | 5.8 | No |
| 22463 | 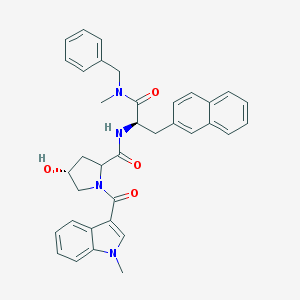 CHEMBL303548 CHEMBL303548 | C36H36N4O4 | 588.708 | 4 / 2 | 4.6 | No |
| 553347 | 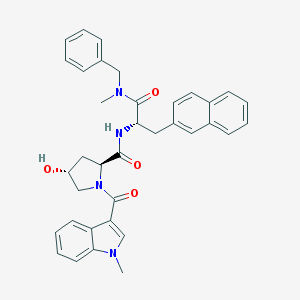 FK 888 FK 888 | C36H36N4O4 | 588.708 | 4 / 2 | 4.6 | No |
| 22467 | 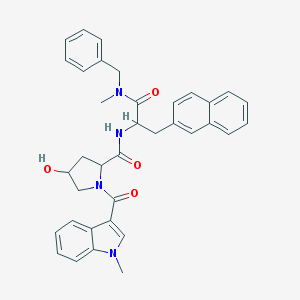 L001421 L001421 | C36H36N4O4 | 588.708 | 4 / 2 | 4.6 | No |
| 23119 | 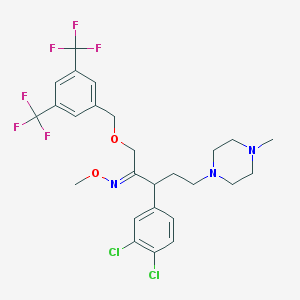 CHEMBL310019 CHEMBL310019 | C26H29Cl2F6N3O2 | 600.427 | 11 / 0 | 6.8 | No |
| 558002 | 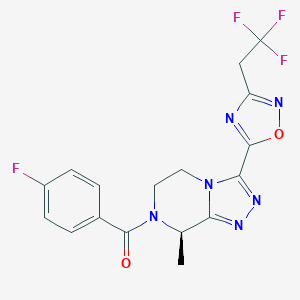 SCHEMBL16115678 SCHEMBL16115678 | C17H14F4N6O2 | 410.333 | 10 / 0 | 1.9 | Yes |
| 548155 | 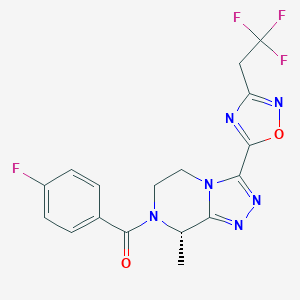 CHEMBL3898651 CHEMBL3898651 | C17H14F4N6O2 | 410.333 | 10 / 0 | 1.9 | Yes |
| 23614 | 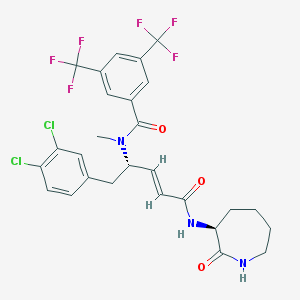 CHEMBL115633 CHEMBL115633 | C27H25Cl2F6N3O3 | 624.405 | 9 / 2 | 6.2 | No |
| 23617 | 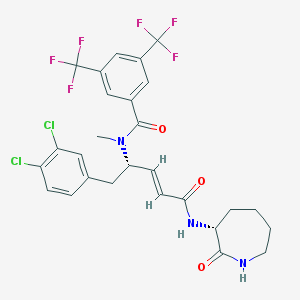 CHEMBL119126 CHEMBL119126 | C27H25Cl2F6N3O3 | 624.405 | 9 / 2 | 6.2 | No |
| 23619 | 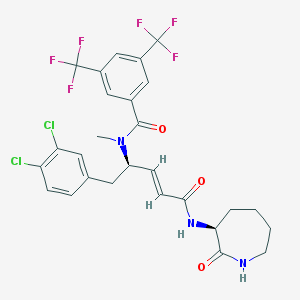 CHEMBL332793 CHEMBL332793 | C27H25Cl2F6N3O3 | 624.405 | 9 / 2 | 6.2 | No |
| 23621 | 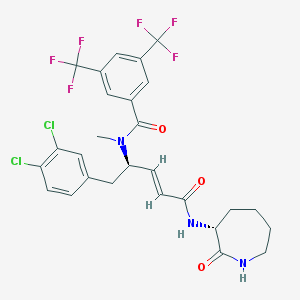 UNII-G9BL4978MZ UNII-G9BL4978MZ | C27H25Cl2F6N3O3 | 624.405 | 9 / 2 | 6.2 | No |
| 23631 | 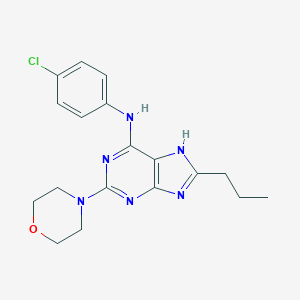 CHEMBL318254 CHEMBL318254 | C18H21ClN6O | 372.857 | 6 / 2 | 3.9 | Yes |
| 24253 | 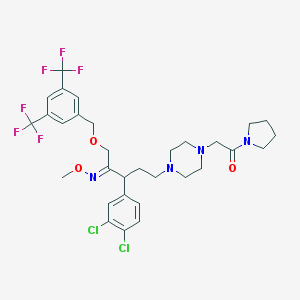 CHEMBL310703 CHEMBL310703 | C31H36Cl2F6N4O3 | 697.544 | 12 / 0 | 6.8 | No |
| 24270 | 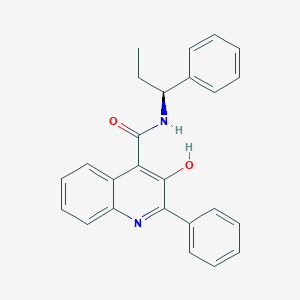 Talnetant Talnetant | C25H22N2O2 | 382.463 | 3 / 2 | 5.7 | No |
| 24275 | 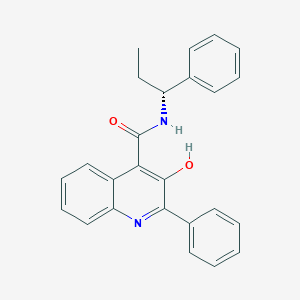 CHEMBL430118 CHEMBL430118 | C25H22N2O2 | 382.463 | 3 / 2 | 5.7 | No |
| 24277 | 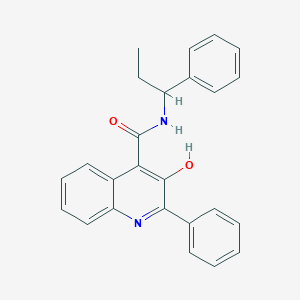 SB 223412 SB 223412 | C25H22N2O2 | 382.463 | 3 / 2 | 5.7 | No |
| 24552 | 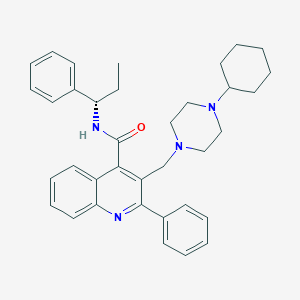 CHEMBL33868 CHEMBL33868 | C36H42N4O | 546.759 | 4 / 1 | 7.0 | No |
| 24756 | 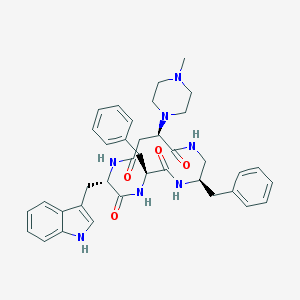 CHEMBL439377 CHEMBL439377 | C38H45N7O4 | 663.823 | 6 / 5 | 3.3 | No |
| 25046 | 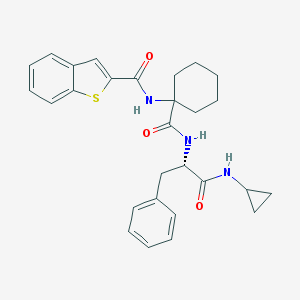 CHEMBL391710 CHEMBL391710 | C28H31N3O3S | 489.634 | 4 / 3 | 5.4 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417