

You can:
| Name | Follicle-stimulating hormone receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | FSHR |
| Synonym | Follitropin receptor FSH receptor FSH-R LGR1 |
| Disease | Female infertility African trypanosomiasis Contraception Ovarian cancer |
| Length | 695 |
| Amino acid sequence | MALLLVSLLAFLSLGSGCHHRICHCSNRVFLCQESKVTEIPSDLPRNAIELRFVLTKLRVIQKGAFSGFGDLEKIEISQNDVLEVIEADVFSNLPKLHEIRIEKANNLLYINPEAFQNLPNLQYLLISNTGIKHLPDVHKIHSLQKVLLDIQDNINIHTIERNSFVGLSFESVILWLNKNGIQEIHNCAFNGTQLDELNLSDNNNLEELPNDVFHGASGPVILDISRTRIHSLPSYGLENLKKLRARSTYNLKKLPTLEKLVALMEASLTYPSHCCAFANWRRQISELHPICNKSILRQEVDYMTQARGQRSSLAEDNESSYSRGFDMTYTEFDYDLCNEVVDVTCSPKPDAFNPCEDIMGYNILRVLIWFISILAITGNIIVLVILTTSQYKLTVPRFLMCNLAFADLCIGIYLLLIASVDIHTKSQYHNYAIDWQTGAGCDAAGFFTVFASELSVYTLTAITLERWHTITHAMQLDCKVQLRHAASVMVMGWIFAFAAALFPIFGISSYMKVSICLPMDIDSPLSQLYVMSLLVLNVLAFVVICGCYIHIYLTVRNPNIVSSSSDTRIAKRMAMLIFTDFLCMAPISFFAISASLKVPLITVSKAKILLVLFHPINSCANPFLYAIFTKNFRRDFFILLSKCGCYEMQAQIYRTETSSTVHNTHPRNGHCSSAPRVTNGSTYILVPLSHLAQN |
| UniProt | P23945 |
| Protein Data Bank | 4mqw |
| GPCR-HGmod model | N/A |
| 3D structure model | This structure is from PDB ID 4mqw. |
| BioLiP | BL0274372 |
| Therapeutic Target Database | T68334 |
| ChEMBL | CHEMBL2024 |
| IUPHAR | N/A |
| DrugBank | BE0000387 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 2132 | 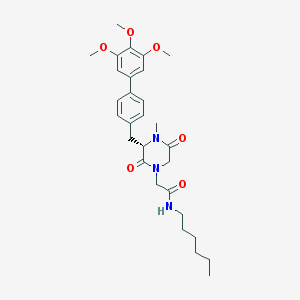 CHEMBL300022 CHEMBL300022 | C29H39N3O6 | 525.646 | 6 / 1 | 4.3 | No |
| 521571 | 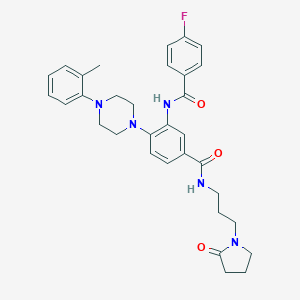 CHEMBL3809148 CHEMBL3809148 | C32H36FN5O3 | 557.67 | 6 / 2 | 4.1 | No |
| 12040 | 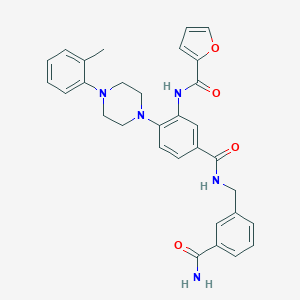 CHEMBL3263685 CHEMBL3263685 | C31H31N5O4 | 537.62 | 6 / 3 | 3.9 | No |
| 14799 | 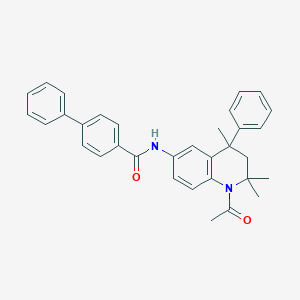 CHEMBL192135 CHEMBL192135 | C33H32N2O2 | 488.631 | 2 / 1 | 6.8 | No |
| 16063 |  CHEMBL3263663 CHEMBL3263663 | C31H37N5O4 | 543.668 | 6 / 2 | 3.8 | No |
| 20401 |  CHEMBL436481 CHEMBL436481 | C34H34N2O2 | 502.658 | 2 / 1 | 7.1 | No |
| 21121 |  CHEMBL370917 CHEMBL370917 | C28H31NO2 | 413.561 | 2 / 0 | 6.2 | No |
| 29083 |  CHEMBL363164 CHEMBL363164 | C33H32N2O3 | 504.63 | 3 / 2 | 6.4 | No |
| 33003 | 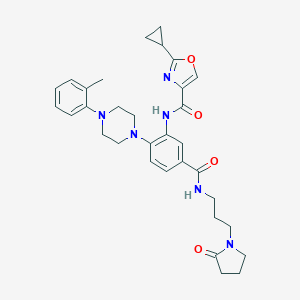 CHEMBL3263667 CHEMBL3263667 | C32H38N6O4 | 570.694 | 7 / 2 | 3.4 | No |
| 43423 | 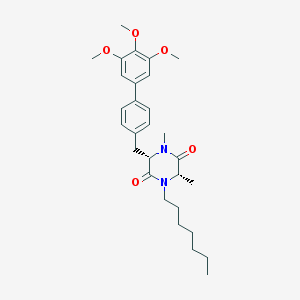 CHEMBL279759 CHEMBL279759 | C29H40N2O5 | 496.648 | 5 / 0 | 5.9 | No |
| 49717 | 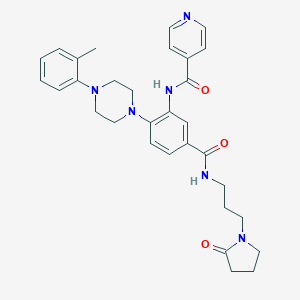 CHEMBL3263673 CHEMBL3263673 | C31H36N6O3 | 540.668 | 6 / 2 | 3.0 | No |
| 50941 | 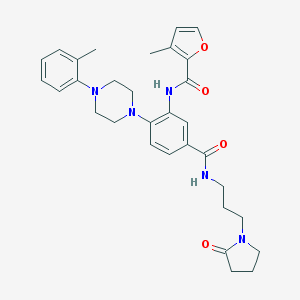 CHEMBL3263664 CHEMBL3263664 | C31H37N5O4 | 543.668 | 6 / 2 | 3.8 | No |
| 57384 |  CHEMBL1651828 CHEMBL1651828 | C29H38N12O4S2 | 682.823 | 14 / 4 | 3.7 | No |
| 57469 |  CHEMBL47059 CHEMBL47059 | C34H49N3O3 | 547.784 | 3 / 0 | 7.0 | No |
| 523307 |  CHEMBL3808949 CHEMBL3808949 | C30H36N6O4 | 544.656 | 6 / 4 | 3.7 | No |
| 63245 |  CHEMBL3263666 CHEMBL3263666 | C30H36N6O4 | 544.656 | 7 / 2 | 3.2 | No |
| 64117 | 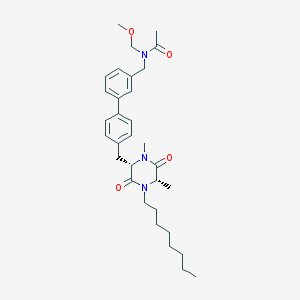 CHEMBL46216 CHEMBL46216 | C32H45N3O4 | 535.729 | 4 / 0 | 5.7 | No |
| 69777 | 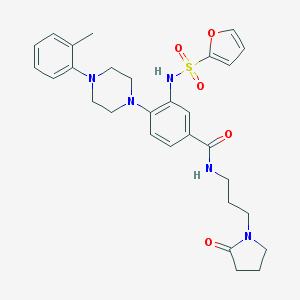 CHEMBL3263662 CHEMBL3263662 | C29H35N5O5S | 565.689 | 8 / 2 | 3.1 | No |
| 70138 | 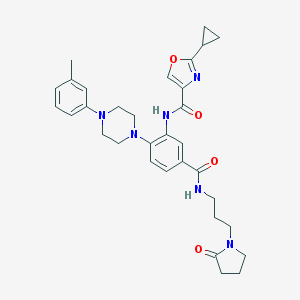 CHEMBL3263676 CHEMBL3263676 | C32H38N6O4 | 570.694 | 7 / 2 | 3.4 | No |
| 520031 | 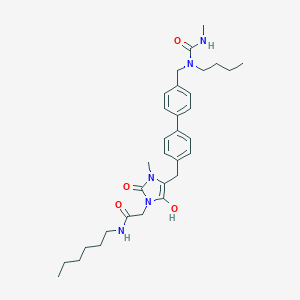 BDBM50142655 BDBM50142655 | C32H45N5O4 | 563.743 | 4 / 3 | 5.0 | No |
| 78873 | 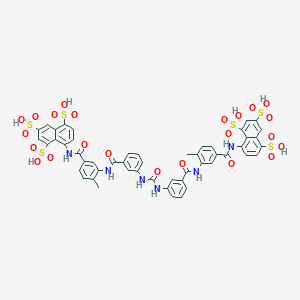 suramin suramin | C51H40N6O23S6 | 1297.26 | 23 / 12 | 1.5 | No |
| 89872 | 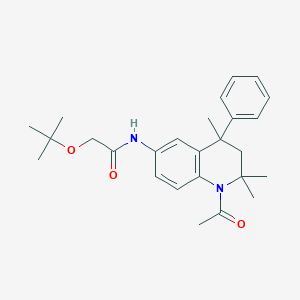 CHEMBL179505 CHEMBL179505 | C26H34N2O3 | 422.569 | 3 / 1 | 4.3 | Yes |
| 91753 | 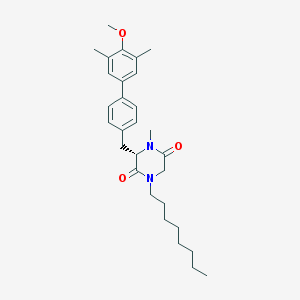 CHEMBL417003 CHEMBL417003 | C29H40N2O3 | 464.65 | 3 / 0 | 6.8 | No |
| 92358 | 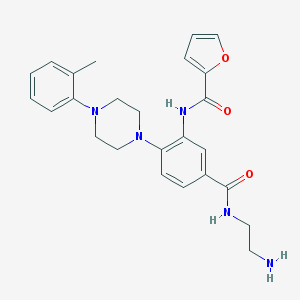 CHEMBL3263684 CHEMBL3263684 | C25H29N5O3 | 447.539 | 6 / 3 | 2.6 | Yes |
| 95970 | 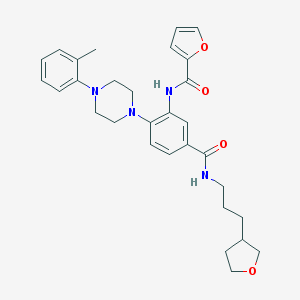 CHEMBL3263687 CHEMBL3263687 | C30H36N4O4 | 516.642 | 6 / 2 | 4.5 | No |
| 101682 | 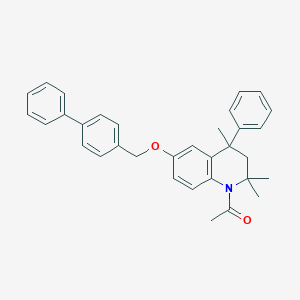 CHEMBL372649 CHEMBL372649 | C33H33NO2 | 475.632 | 2 / 0 | 7.4 | No |
| 105558 | 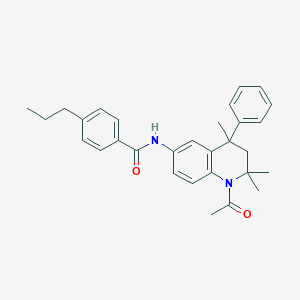 CHEMBL360981 CHEMBL360981 | C30H34N2O2 | 454.614 | 2 / 1 | 6.5 | No |
| 109890 | 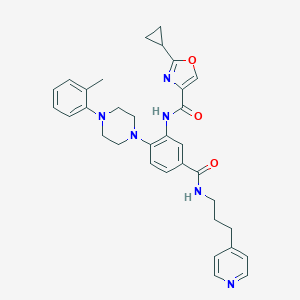 CHEMBL3263688 CHEMBL3263688 | C33H36N6O3 | 564.69 | 7 / 2 | 4.7 | No |
| 524798 | 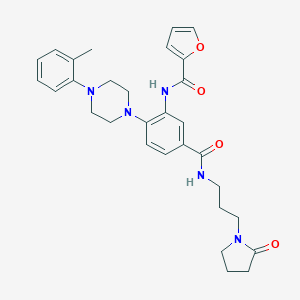 CHEMBL3263694 CHEMBL3263694 | C30H35N5O4 | 529.641 | 6 / 2 | 3.4 | No |
| 118598 | 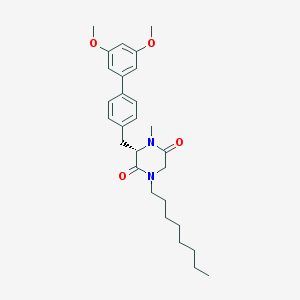 CHEMBL45782 CHEMBL45782 | C28H38N2O4 | 466.622 | 4 / 0 | 6.0 | No |
| 119926 | 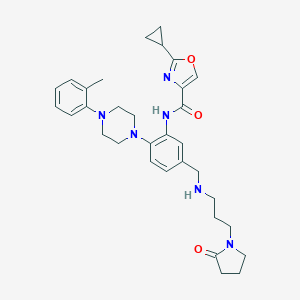 CHEMBL3263693 CHEMBL3263693 | C32H40N6O3 | 556.711 | 7 / 2 | 3.5 | No |
| 120884 | 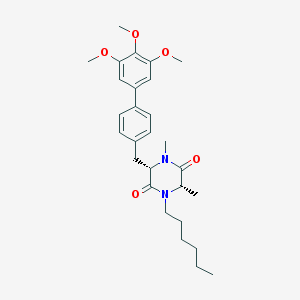 CHEMBL47500 CHEMBL47500 | C28H38N2O5 | 482.621 | 5 / 0 | 5.3 | No |
| 121918 | 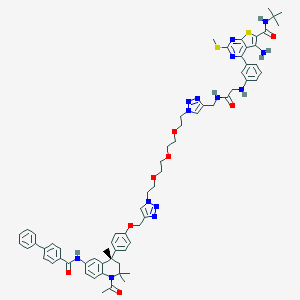 CHEMBL1651822 CHEMBL1651822 | C67H76N14O8S2 | 1269.56 | 18 / 5 | 8.4 | No |
| 121920 | 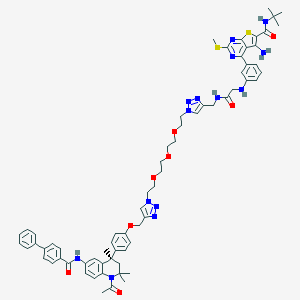 CHEMBL1651825 CHEMBL1651825 | C67H76N14O8S2 | 1269.56 | 18 / 5 | 8.4 | No |
| 128729 | 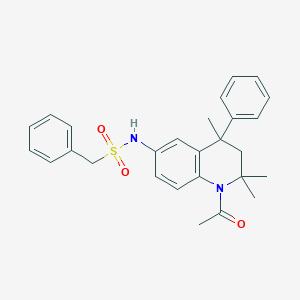 CHEMBL371595 CHEMBL371595 | C27H30N2O3S | 462.608 | 4 / 1 | 4.7 | Yes |
| 134260 | 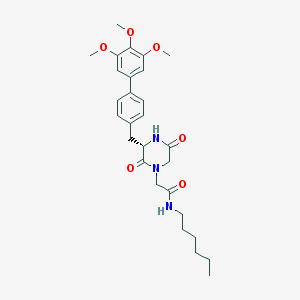 CHEMBL43789 CHEMBL43789 | C28H37N3O6 | 511.619 | 6 / 2 | 4.1 | No |
| 139406 | 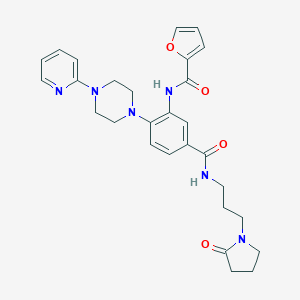 CHEMBL3263678 CHEMBL3263678 | C28H32N6O4 | 516.602 | 7 / 2 | 2.3 | No |
| 140323 | 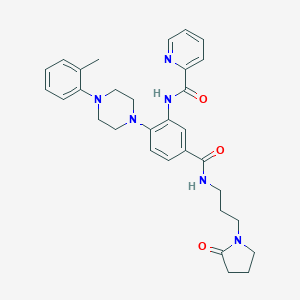 CHEMBL3263671 CHEMBL3263671 | C31H36N6O3 | 540.668 | 6 / 2 | 3.3 | No |
| 145222 | 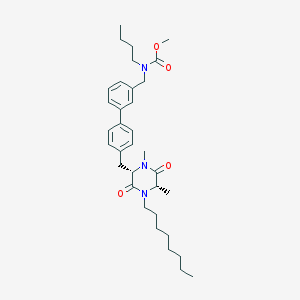 CHEMBL433263 CHEMBL433263 | C34H49N3O4 | 563.783 | 4 / 0 | 7.4 | No |
| 146152 | 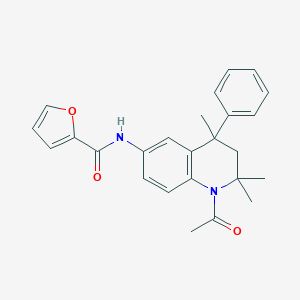 CHEMBL361167 CHEMBL361167 | C25H26N2O3 | 402.494 | 3 / 1 | 4.5 | Yes |
| 149654 | 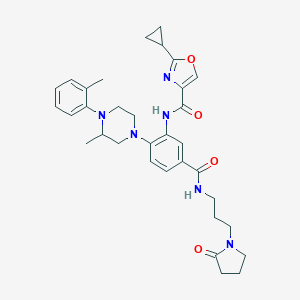 CHEMBL3263681 CHEMBL3263681 | C33H40N6O4 | 584.721 | 7 / 2 | 3.8 | No |
| 163274 | 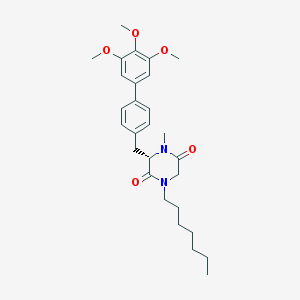 CHEMBL298686 CHEMBL298686 | C28H38N2O5 | 482.621 | 5 / 0 | 5.5 | No |
| 165071 | 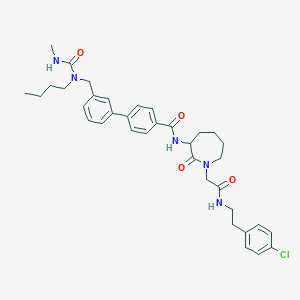 CHEMBL50556 CHEMBL50556 | C36H44ClN5O4 | 646.229 | 4 / 3 | 5.5 | No |
| 170438 | 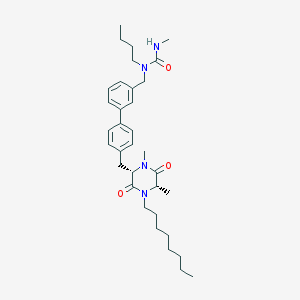 CHEMBL295895 CHEMBL295895 | C34H50N4O3 | 562.799 | 3 / 1 | 6.9 | No |
| 171566 | 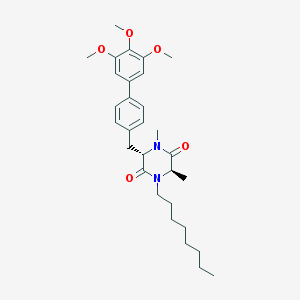 CHEMBL49308 CHEMBL49308 | C30H42N2O5 | 510.675 | 5 / 0 | 6.4 | No |
| 171567 | 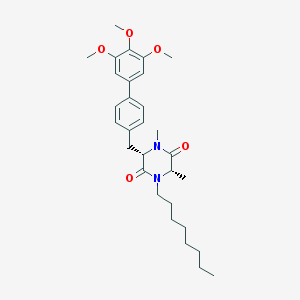 CHEMBL49624 CHEMBL49624 | C30H42N2O5 | 510.675 | 5 / 0 | 6.4 | No |
| 171568 | 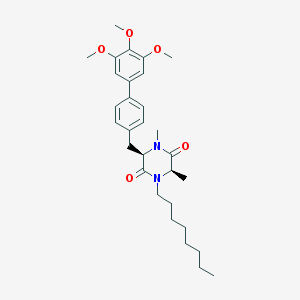 CHEMBL49540 CHEMBL49540 | C30H42N2O5 | 510.675 | 5 / 0 | 6.4 | No |
| 171569 | 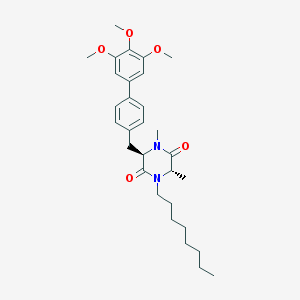 CHEMBL296449 CHEMBL296449 | C30H42N2O5 | 510.675 | 5 / 0 | 6.4 | No |
| 179481 | 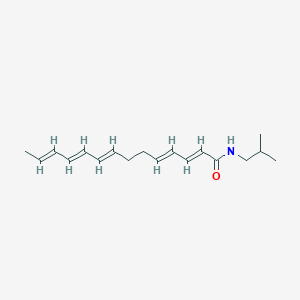 N-Isobutyl-2,4,8,10,12-tetradecapentaenamide N-Isobutyl-2,4,8,10,12-tetradecapentaenamide | C18H27NO | 273.42 | 1 / 1 | 4.9 | Yes |
| 186114 | 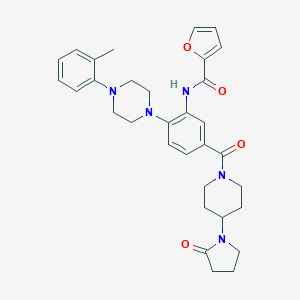 CHEMBL3263682 CHEMBL3263682 | C32H37N5O4 | 555.679 | 6 / 1 | 3.8 | No |
| 189295 | 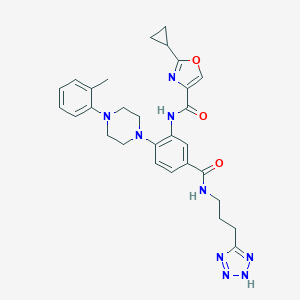 CHEMBL3263691 CHEMBL3263691 | C29H33N9O3 | 555.643 | 9 / 3 | 3.3 | No |
| 191641 | 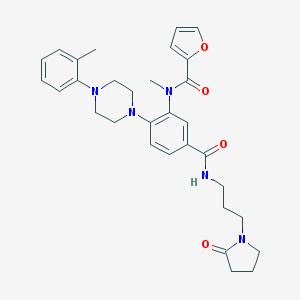 CHEMBL3263661 CHEMBL3263661 | C31H37N5O4 | 543.668 | 6 / 1 | 3.6 | No |
| 193399 |  CHEMBL1651821 CHEMBL1651821 | C65H72N14O7S2 | 1225.5 | 17 / 5 | 8.5 | No |
| 193400 |  CHEMBL1651824 CHEMBL1651824 | C65H72N14O7S2 | 1225.5 | 17 / 5 | 8.5 | No |
| 194991 |  CHEMBL195321 CHEMBL195321 | C30H34N2O3 | 470.613 | 3 / 1 | 5.5 | No |
| 195794 |  CHEMBL3263670 CHEMBL3263670 | C32H37N5O3 | 539.68 | 5 / 2 | 4.0 | No |
| 196108 |  CHEMBL49608 CHEMBL49608 | C32H46N4O3 | 534.745 | 3 / 1 | 5.8 | No |
| 202967 |  CHEMBL3263660 CHEMBL3263660 | C30H35N5O4 | 529.641 | 6 / 2 | 3.1 | No |
| 205966 |  CHEMBL3263665 CHEMBL3263665 | C31H38N6O3 | 542.684 | 5 / 2 | 3.3 | No |
| 206762 |  CHEMBL1651831 CHEMBL1651831 | C69H80N14O9S2 | 1313.61 | 19 / 5 | 8.2 | No |
| 206764 |  CHEMBL1651826 CHEMBL1651826 | C69H80N14O9S2 | 1313.61 | 19 / 5 | 8.2 | No |
| 217816 |  CHEMBL192868 CHEMBL192868 | C34H34N2O3 | 518.657 | 3 / 1 | 6.7 | No |
| 222019 |  CHEMBL3263686 CHEMBL3263686 | C30H36N4O3 | 500.643 | 5 / 2 | 6.1 | No |
| 229442 |  CHEMBL372506 CHEMBL372506 | C27H29N3O2 | 427.548 | 2 / 2 | 4.9 | Yes |
| 237342 | 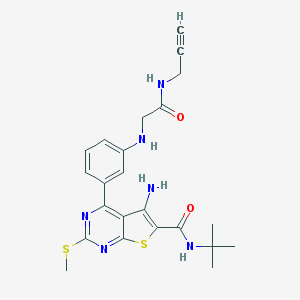 CHEMBL1651721 CHEMBL1651721 | C23H26N6O2S2 | 482.621 | 8 / 4 | 4.0 | Yes |
| 242785 | 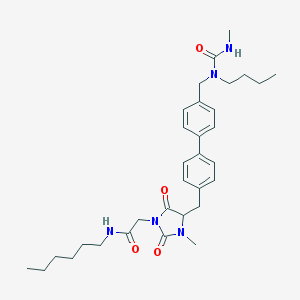 CID 44292945 CID 44292945 | C32H45N5O4 | 563.743 | 4 / 2 | 5.0 | No |
| 243489 | 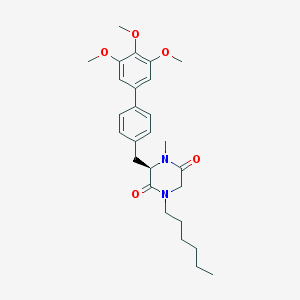 CHEMBL300634 CHEMBL300634 | C27H36N2O5 | 468.594 | 5 / 0 | 4.9 | Yes |
| 243490 | 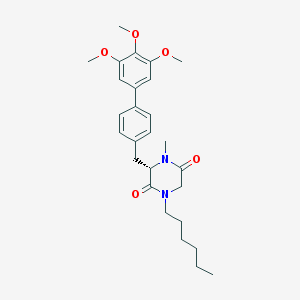 CHEMBL50278 CHEMBL50278 | C27H36N2O5 | 468.594 | 5 / 0 | 4.9 | Yes |
| 244764 | 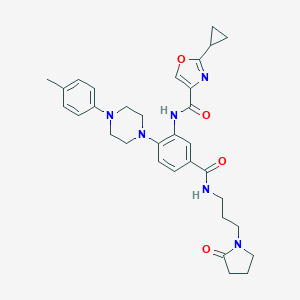 CHEMBL3263677 CHEMBL3263677 | C32H38N6O4 | 570.694 | 7 / 2 | 3.4 | No |
| 245851 | 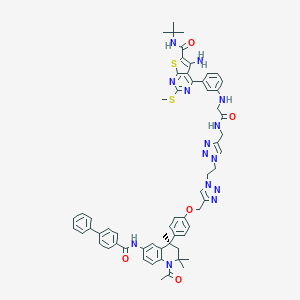 CHEMBL1651723 CHEMBL1651723 | C61H64N14O5S2 | 1137.4 | 15 / 5 | 8.8 | No |
| 245853 | 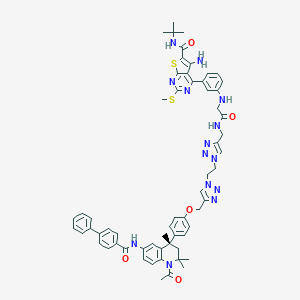 CHEMBL1651829 CHEMBL1651829 | C61H64N14O5S2 | 1137.4 | 15 / 5 | 8.8 | No |
| 247438 | 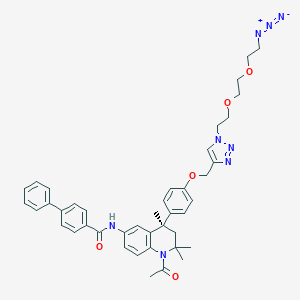 CHEMBL1651827 CHEMBL1651827 | C42H46N8O5 | 742.881 | 9 / 1 | 6.6 | No |
| 250807 | 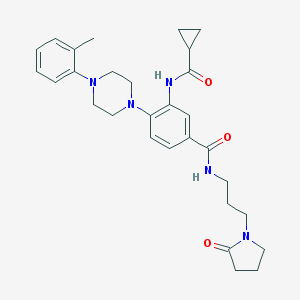 CHEMBL3263669 CHEMBL3263669 | C29H37N5O3 | 503.647 | 5 / 2 | 2.8 | No |
| 251595 | 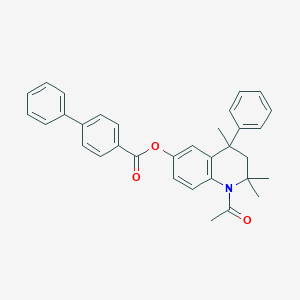 CHEMBL192334 CHEMBL192334 | C33H31NO3 | 489.615 | 3 / 0 | 7.3 | No |
| 257274 | 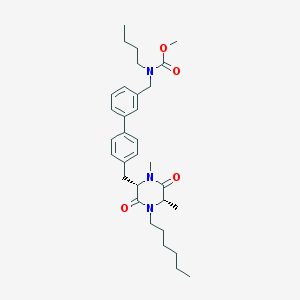 CHEMBL48375 CHEMBL48375 | C32H45N3O4 | 535.729 | 4 / 0 | 6.4 | No |
| 259382 | 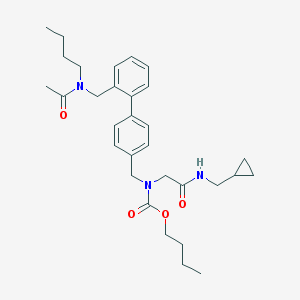 CHEMBL47120 CHEMBL47120 | C31H43N3O4 | 521.702 | 4 / 1 | 5.1 | No |
| 266896 | 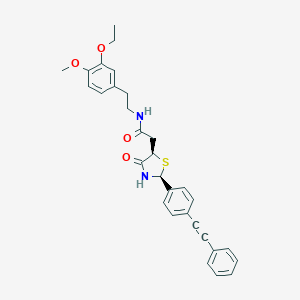 CHEMBL2216804 CHEMBL2216804 | C30H30N2O4S | 514.64 | 5 / 2 | 5.2 | No |
| 268222 | 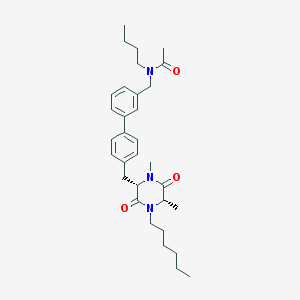 CHEMBL44663 CHEMBL44663 | C32H45N3O3 | 519.73 | 3 / 0 | 6.0 | No |
| 278854 | 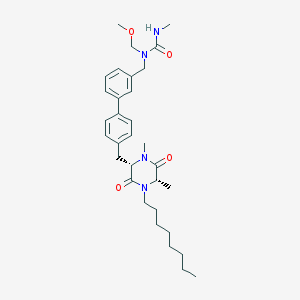 CHEMBL48030 CHEMBL48030 | C32H46N4O4 | 550.744 | 4 / 1 | 5.6 | No |
| 281917 | 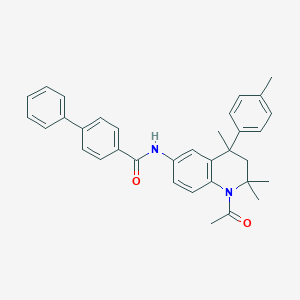 CHEMBL195347 CHEMBL195347 | C34H34N2O2 | 502.658 | 2 / 1 | 7.1 | No |
| 285662 | 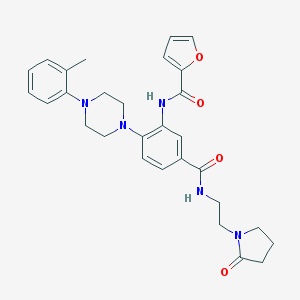 CHEMBL3263683 CHEMBL3263683 | C29H33N5O4 | 515.614 | 6 / 2 | 3.1 | No |
| 299443 | 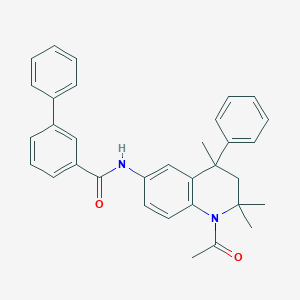 CHEMBL365545 CHEMBL365545 | C33H32N2O2 | 488.631 | 2 / 1 | 6.8 | No |
| 300434 | 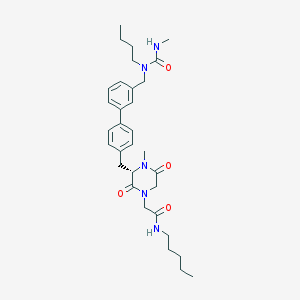 CHEMBL45770 CHEMBL45770 | C32H45N5O4 | 563.743 | 4 / 2 | 4.2 | No |
| 530016 | 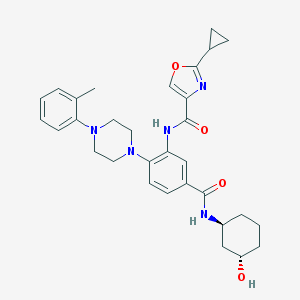 CHEMBL3809644 CHEMBL3809644 | C31H37N5O4 | 543.668 | 7 / 3 | 4.1 | No |
| 304815 | 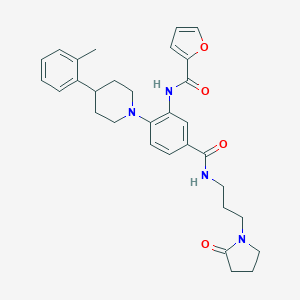 CHEMBL3263680 CHEMBL3263680 | C31H36N4O4 | 528.653 | 5 / 2 | 4.1 | No |
| 305015 | 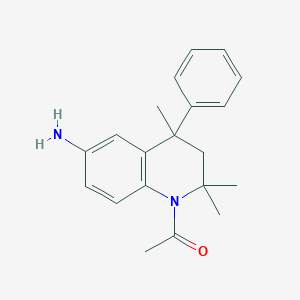 CHEMBL195534 CHEMBL195534 | C20H24N2O | 308.425 | 2 / 1 | 3.6 | Yes |
| 316239 | 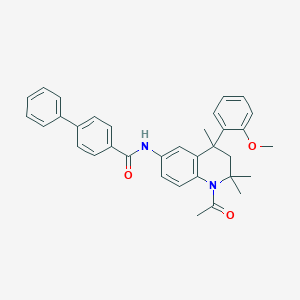 CHEMBL426824 CHEMBL426824 | C34H34N2O3 | 518.657 | 3 / 1 | 6.7 | No |
| 316244 | 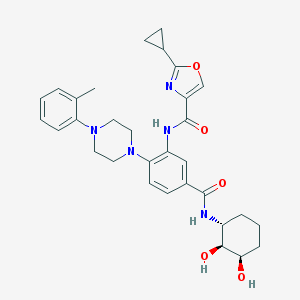 CHEMBL3263692 CHEMBL3263692 | C31H37N5O5 | 559.667 | 8 / 4 | 3.1 | No |
| 323314 | 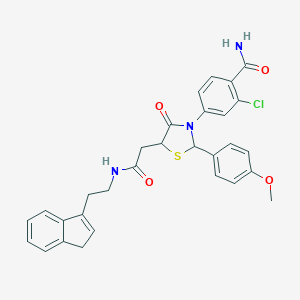 CHEMBL524348 CHEMBL524348 | C30H28ClN3O4S | 562.081 | 5 / 2 | 4.4 | No |
| 335462 | 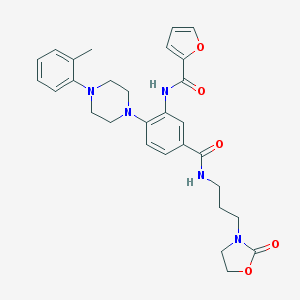 CHEMBL3263689 CHEMBL3263689 | C29H33N5O5 | 531.613 | 7 / 2 | 3.5 | No |
| 337501 | 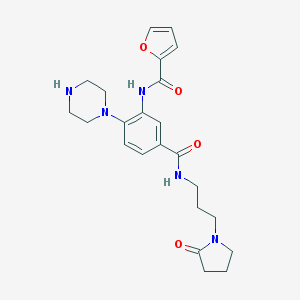 CHEMBL3263679 CHEMBL3263679 | C23H29N5O4 | 439.516 | 6 / 3 | 0.8 | Yes |
| 347120 | 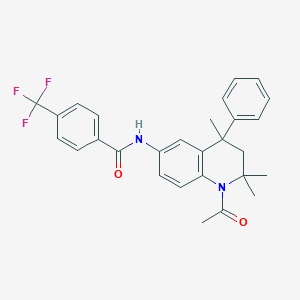 CHEMBL361847 CHEMBL361847 | C28H27F3N2O2 | 480.531 | 5 / 1 | 6.0 | No |
| 347543 | 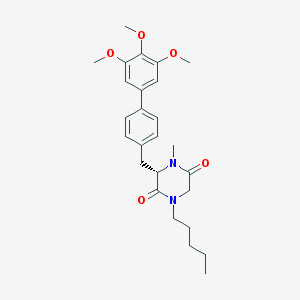 CHEMBL432883 CHEMBL432883 | C26H34N2O5 | 454.567 | 5 / 0 | 4.4 | Yes |
| 350593 | 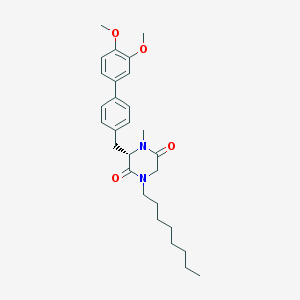 CHEMBL47420 CHEMBL47420 | C28H38N2O4 | 466.622 | 4 / 0 | 6.0 | No |
| 351711 | 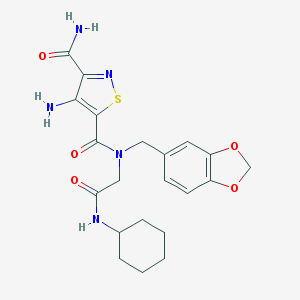 AC1MLEST AC1MLEST | C21H25N5O5S | 459.521 | 8 / 3 | 2.3 | Yes |
| 355266 | 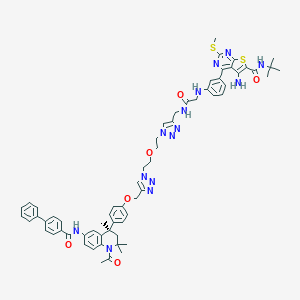 CHEMBL1651823 CHEMBL1651823 | C63H68N14O6S2 | 1181.45 | 16 / 5 | 8.7 | No |
| 355268 | 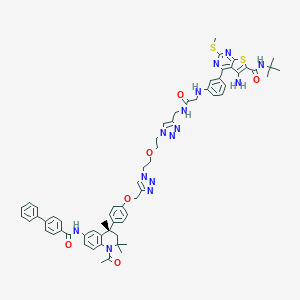 CHEMBL1651830 CHEMBL1651830 | C63H68N14O6S2 | 1181.45 | 16 / 5 | 8.7 | No |
| 355775 | 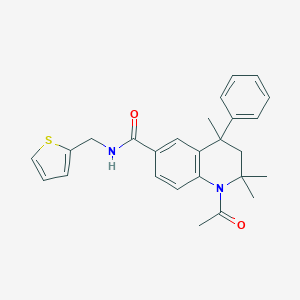 CHEMBL364094 CHEMBL364094 | C26H28N2O2S | 432.582 | 3 / 1 | 4.8 | Yes |
| 367991 | 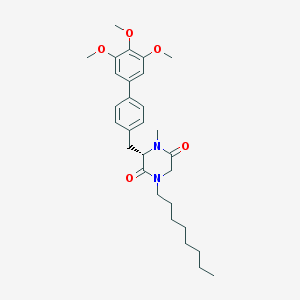 CHEMBL45976 CHEMBL45976 | C29H40N2O5 | 496.648 | 5 / 0 | 6.0 | No |
| 369790 | 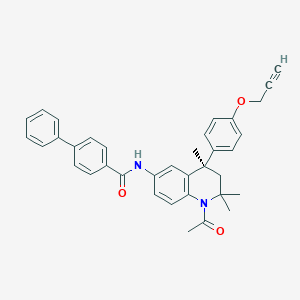 CHEMBL1651722 CHEMBL1651722 | C36H34N2O3 | 542.679 | 3 / 1 | 6.8 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417