

You can:
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 47 | 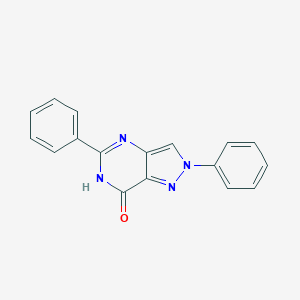 CHEMBL574416 CHEMBL574416 | C17H12N4O | 288.31 | 3 / 1 | 2.7 | Yes |
| 50 | 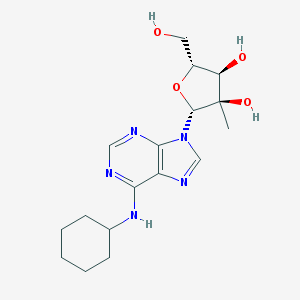 CHEMBL2113609 CHEMBL2113609 | C17H25N5O4 | 363.418 | 8 / 4 | 0.8 | Yes |
| 70 | 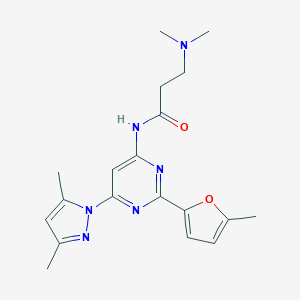 Pyrimidin-4-ylacetamide, 26 Pyrimidin-4-ylacetamide, 26 | C19H24N6O2 | 368.441 | 6 / 1 | 2.0 | Yes |
| 157 | 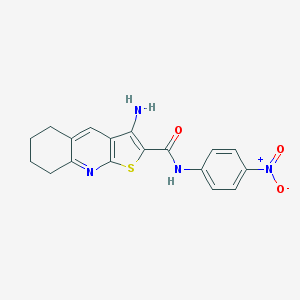 CHEMBL601208 CHEMBL601208 | C18H16N4O3S | 368.411 | 6 / 2 | 4.3 | Yes |
| 188 | 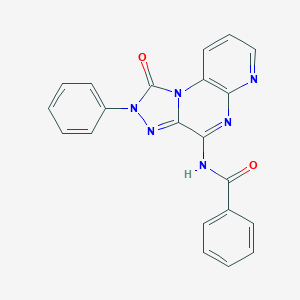 CHEMBL473650 CHEMBL473650 | C21H14N6O2 | 382.383 | 5 / 1 | 2.8 | Yes |
| 441675 | 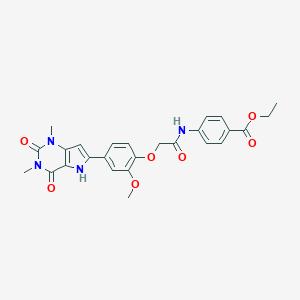 CHEMBL201449 CHEMBL201449 | C26H26N4O7 | 506.515 | 7 / 2 | 2.7 | No |
| 236 | 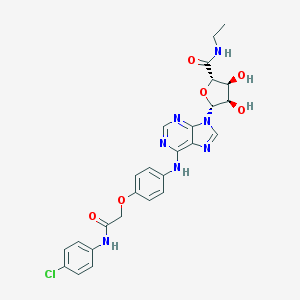 CHEMBL437168 CHEMBL437168 | C26H26ClN7O6 | 567.987 | 10 / 5 | 2.2 | No |
| 366 | 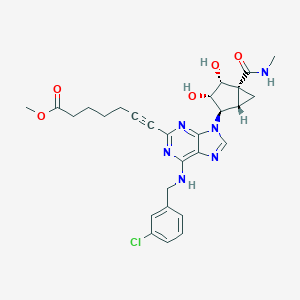 CHEMBL573125 CHEMBL573125 | C28H31ClN6O5 | 567.043 | 9 / 4 | 2.5 | No |
| 441688 | 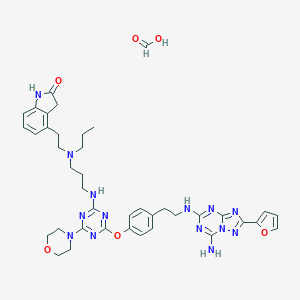 CHEMBL3394262 CHEMBL3394262 | C40H48N14O6 | 820.916 | 18 / 5 | N/A | No |
| 478 | 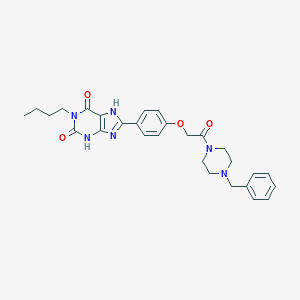 CHEMBL38304 CHEMBL38304 | C28H32N6O4 | 516.602 | 6 / 2 | 3.2 | No |
| 670 | 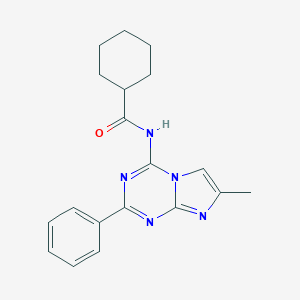 CHEMBL343256 CHEMBL343256 | C19H21N5O | 335.411 | 4 / 1 | 4.5 | Yes |
| 689 | 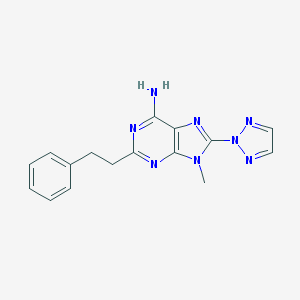 CHEMBL196259 CHEMBL196259 | C16H16N8 | 320.36 | 6 / 1 | 2.3 | Yes |
| 521468 | 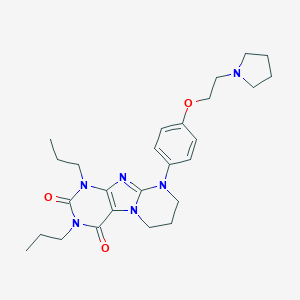 CHEMBL3827623 CHEMBL3827623 | C26H36N6O3 | 480.613 | 6 / 0 | 3.9 | Yes |
| 798 | 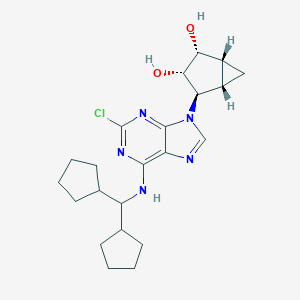 CHEMBL2170801 CHEMBL2170801 | C22H30ClN5O2 | 431.965 | 6 / 3 | 4.2 | Yes |
| 802 | 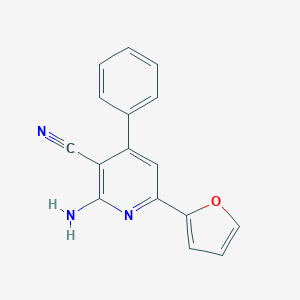 CHEMBL468370 CHEMBL468370 | C16H11N3O | 261.284 | 4 / 1 | 3.2 | Yes |
| 809 | 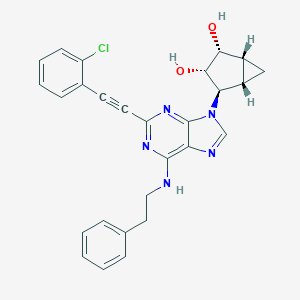 CHEMBL2089347 CHEMBL2089347 | C27H24ClN5O2 | 485.972 | 6 / 3 | 4.2 | Yes |
| 828 | 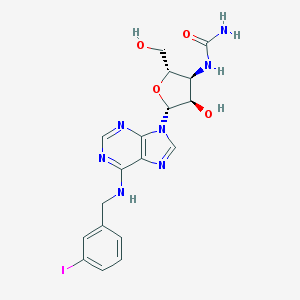 CHEMBL382194 CHEMBL382194 | C18H20IN7O4 | 525.307 | 8 / 5 | 0.4 | No |
| 872 | 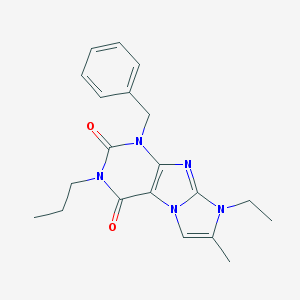 CHEMBL481383 CHEMBL481383 | C20H23N5O2 | 365.437 | 3 / 0 | 3.6 | Yes |
| 896 | 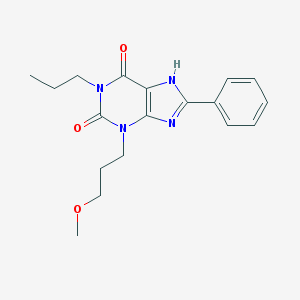 CHEMBL561167 CHEMBL561167 | C18H22N4O3 | 342.399 | 4 / 1 | 3.3 | Yes |
| 935 | 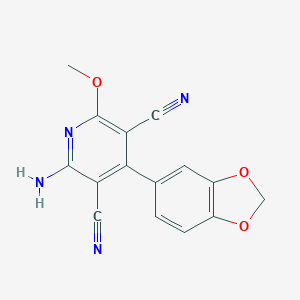 2-amino-4-(1,3-benzodioxol-5-yl)-6-methoxypyridine-3,5-dicarbonitrile 2-amino-4-(1,3-benzodioxol-5-yl)-6-methoxypyridine-3,5-dicarbonitrile | C15H10N4O3 | 294.27 | 7 / 1 | 2.2 | Yes |
| 993 | 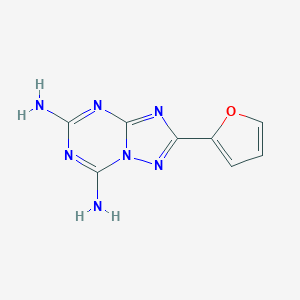 CHEMBL1927632 CHEMBL1927632 | C8H7N7O | 217.192 | 7 / 2 | -0.1 | Yes |
| 998 | 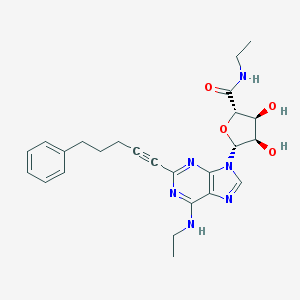 CHEMBL208311 CHEMBL208311 | C25H30N6O4 | 478.553 | 8 / 4 | 2.7 | Yes |
| 1042 | 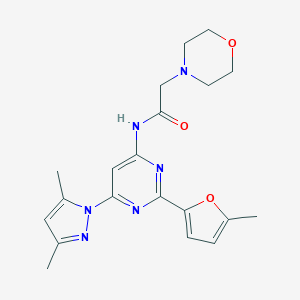 Pyrimidin-4-ylacetamide, 9 Pyrimidin-4-ylacetamide, 9 | C20H24N6O3 | 396.451 | 7 / 1 | 1.7 | Yes |
| 1136 | 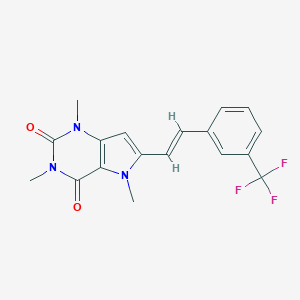 CHEMBL2313295 CHEMBL2313295 | C18H16F3N3O2 | 363.34 | 5 / 0 | 3.1 | Yes |
| 1201 | 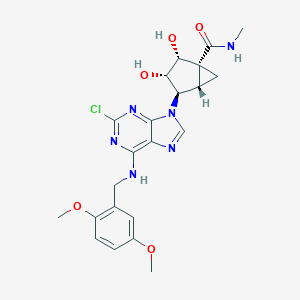 CHEMBL178846 CHEMBL178846 | C22H25ClN6O5 | 488.929 | 9 / 4 | 1.3 | Yes |
| 553256 | 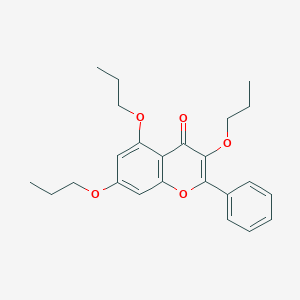 MRS-1042 MRS-1042 | C24H28O5 | 396.483 | 5 / 0 | 6.0 | No |
| 1449 | 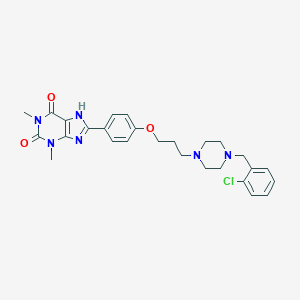 CHEMBL3121728 CHEMBL3121728 | C27H31ClN6O3 | 523.034 | 6 / 1 | 3.6 | No |
| 1466 | 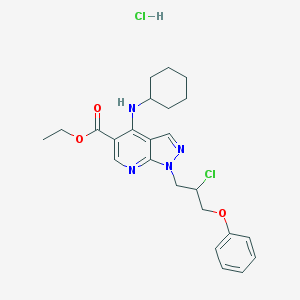 CHEMBL540094 CHEMBL540094 | C24H30Cl2N4O3 | 493.429 | 6 / 2 | N/A | N/A |
| 1477 | 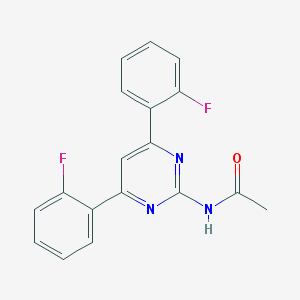 CHEMBL1650163 CHEMBL1650163 | C18H13F2N3O | 325.319 | 5 / 1 | 3.2 | Yes |
| 441727 | 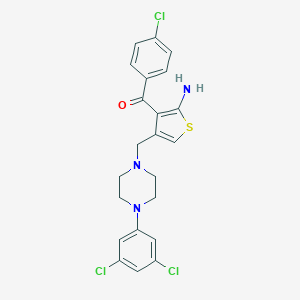 SCHEMBL1497826 SCHEMBL1497826 | C22H20Cl3N3OS | 480.832 | 5 / 1 | 6.5 | No |
| 1560 | 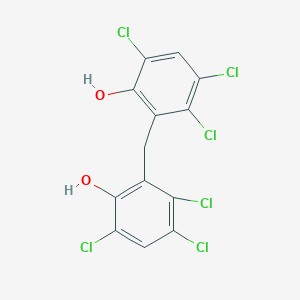 hexachlorophene hexachlorophene | C13H6Cl6O2 | 406.889 | 2 / 2 | 7.5 | No |
| 441734 | 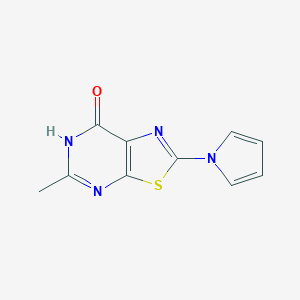 CID 118733042 CID 118733042 | C10H8N4OS | 232.261 | 4 / 1 | 1.1 | Yes |
| 1714 | 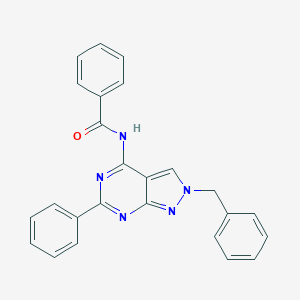 CHEMBL1094143 CHEMBL1094143 | C25H19N5O | 405.461 | 4 / 1 | 4.6 | Yes |
| 1772 | 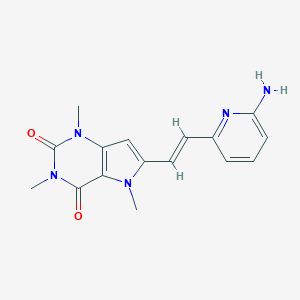 CHEMBL2313274 CHEMBL2313274 | C16H17N5O2 | 311.345 | 4 / 1 | 0.8 | Yes |
| 1817 | 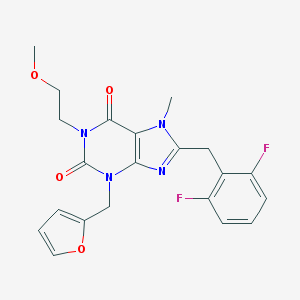 CHEMBL603861 CHEMBL603861 | C21H20F2N4O4 | 430.412 | 7 / 0 | 2.2 | Yes |
| 557350 | 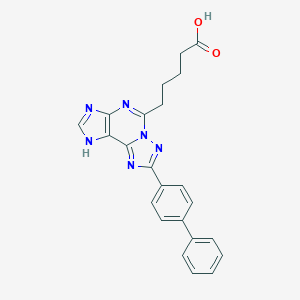 CHEMBL124887 CHEMBL124887 | C23H20N6O2 | 412.453 | 6 / 2 | 3.8 | Yes |
| 1992 | 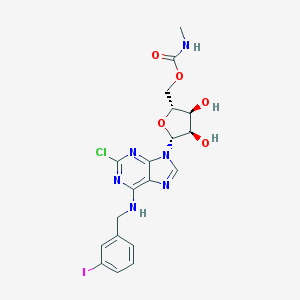 CHEMBL515442 CHEMBL515442 | C19H20ClIN6O5 | 574.76 | 9 / 4 | 1.7 | No |
| 2058 | 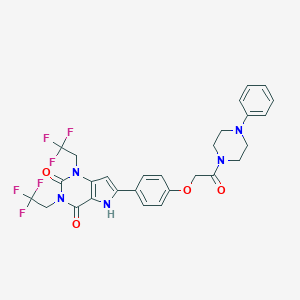 CHEMBL407372 CHEMBL407372 | C28H25F6N5O4 | 609.529 | 11 / 1 | 4.9 | No |
| 2121 | 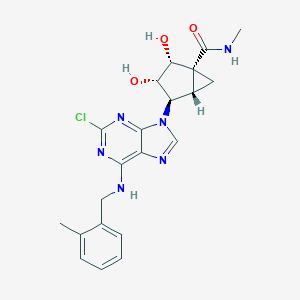 CHEMBL2112158 CHEMBL2112158 | C21H23ClN6O3 | 442.904 | 7 / 4 | 1.7 | Yes |
| 441759 | 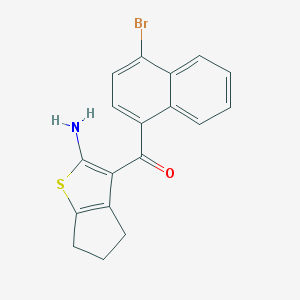 CHEMBL434692 CHEMBL434692 | C18H14BrNOS | 372.28 | 3 / 1 | 5.8 | No |
| 2243 |  CHEMBL259178 CHEMBL259178 | C16H15N5O2 | 309.329 | 6 / 2 | 1.9 | Yes |
| 557360 |  CHEMBL2312720 CHEMBL2312720 | C18H13N3O3 | 319.32 | 4 / 1 | 2.9 | Yes |
| 441764 |  CHEMBL3410319 CHEMBL3410319 | C23H15F4N5O4 | 501.398 | 11 / 0 | 3.9 | No |
| 2308 |  CHEMBL218837 CHEMBL218837 | C19H20ClN7O5 | 461.863 | 9 / 5 | 0.9 | Yes |
| 2317 | 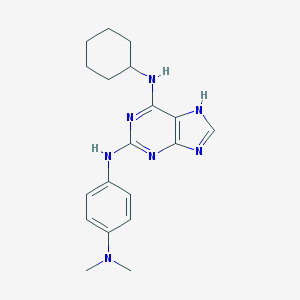 CHEMBL366216 CHEMBL366216 | C19H25N7 | 351.458 | 6 / 3 | 4.2 | Yes |
| 521526 | 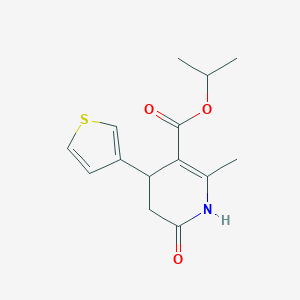 CHEMBL3785142 CHEMBL3785142 | C14H17NO3S | 279.354 | 4 / 1 | 1.8 | Yes |
| 2992 | 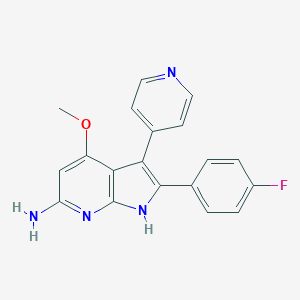 RWJ-68354 RWJ-68354 | C19H15FN4O | 334.354 | 5 / 2 | 3.3 | Yes |
| 3059 | 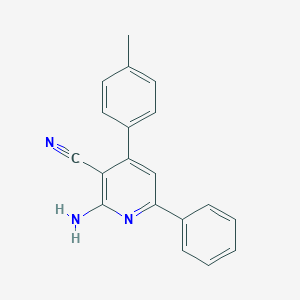 CHEMBL376920 CHEMBL376920 | C19H15N3 | 285.35 | 3 / 1 | 4.4 | Yes |
| 3096 |  CHEMBL131242 CHEMBL131242 | C33H42N8O4 | 614.751 | 10 / 5 | 3.7 | No |
| 3104 |  Pyrimidin-4-ylacetamide, 6 Pyrimidin-4-ylacetamide, 6 | C22H29N7O2 | 423.521 | 7 / 1 | 2.2 | Yes |
| 3185 |  CHEMBL201847 CHEMBL201847 | C26H28N4O4 | 460.534 | 4 / 1 | 3.5 | Yes |
| 3621 |  CHEMBL1927430 CHEMBL1927430 | C19H28N8O5 | 448.484 | 11 / 3 | 1.0 | No |
| 3624 | 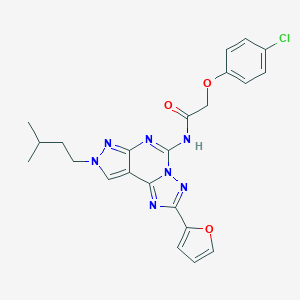 CHEMBL574502 CHEMBL574502 | C23H22ClN7O3 | 479.925 | 7 / 1 | 4.3 | Yes |
| 3673 | 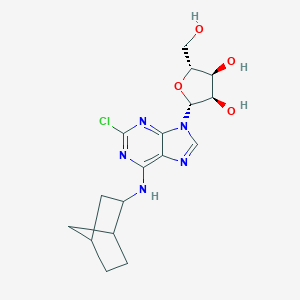 CHEMBL490250 CHEMBL490250 | C17H22ClN5O4 | 395.844 | 8 / 4 | 2.3 | Yes |
| 3789 | 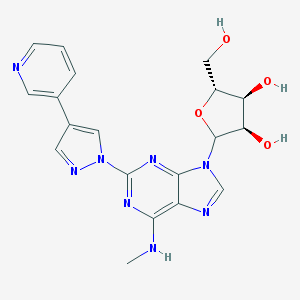 CHEMBL186443 CHEMBL186443 | C19H20N8O4 | 424.421 | 10 / 4 | -0.4 | Yes |
| 3814 | 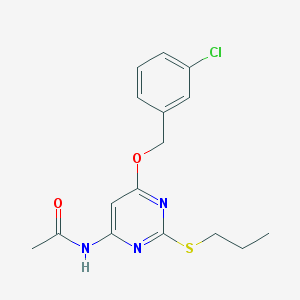 CHEMBL257256 CHEMBL257256 | C16H18ClN3O2S | 351.849 | 5 / 1 | 3.9 | Yes |
| 557391 | 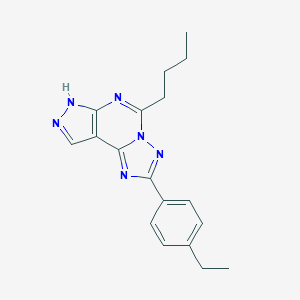 CHEMBL124724 CHEMBL124724 | C18H20N6 | 320.4 | 4 / 1 | 4.3 | Yes |
| 3850 | 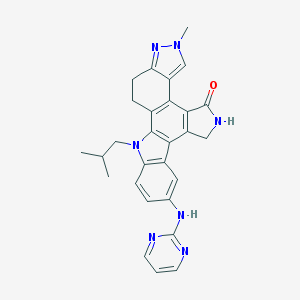 CEP-11981 CEP-11981 | C28H27N7O | 477.572 | 5 / 2 | 3.9 | Yes |
| 3853 | 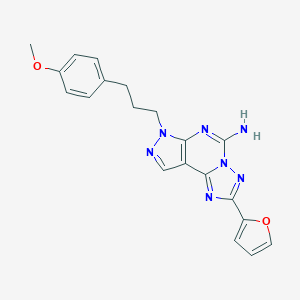 316173-57-6 316173-57-6 | C20H19N7O2 | 389.419 | 7 / 1 | 2.7 | Yes |
| 557393 | 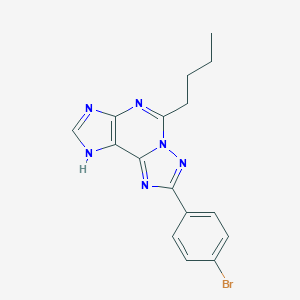 CHEMBL123144 CHEMBL123144 | C16H15BrN6 | 371.242 | 4 / 1 | 4.1 | Yes |
| 3957 | 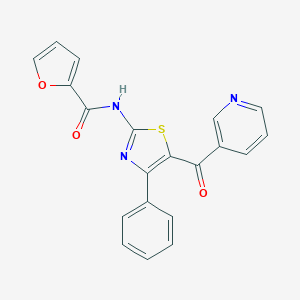 CHEMBL2391832 CHEMBL2391832 | C20H13N3O3S | 375.402 | 6 / 1 | 3.8 | Yes |
| 4041 | 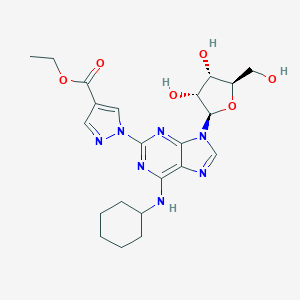 CHEMBL428716 CHEMBL428716 | C22H29N7O6 | 487.517 | 11 / 4 | 1.1 | No |
| 517338 | 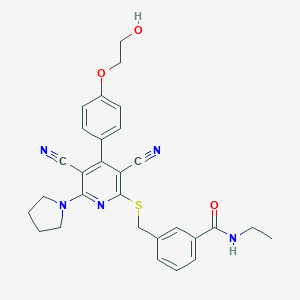 SCHEMBL246621 SCHEMBL246621 | C29H29N5O3S | 527.643 | 8 / 2 | 4.1 | No |
| 4088 | 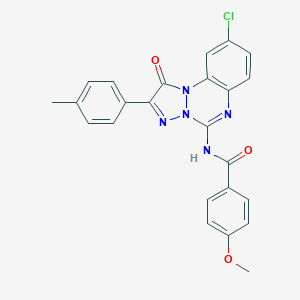 CHEMBL235717 CHEMBL235717 | C24H18ClN5O3 | 459.89 | 5 / 1 | 5.7 | No |
| 557401 | 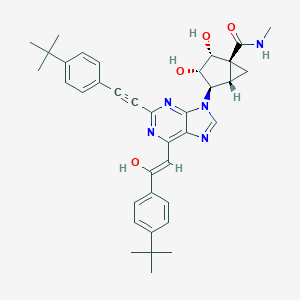 CHEMBL3800558 CHEMBL3800558 | C37H41N5O4 | 619.766 | 7 / 4 | 5.7 | No |
| 4117 | 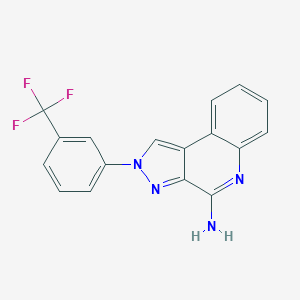 CHEMBL465640 CHEMBL465640 | C17H11F3N4 | 328.298 | 6 / 1 | 4.0 | Yes |
| 4164 | 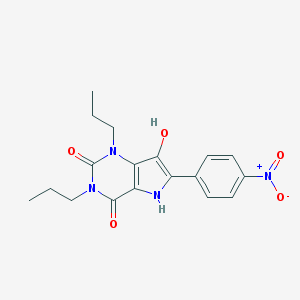 CHEMBL457014 CHEMBL457014 | C18H20N4O5 | 372.381 | 5 / 2 | 2.9 | Yes |
| 4211 | 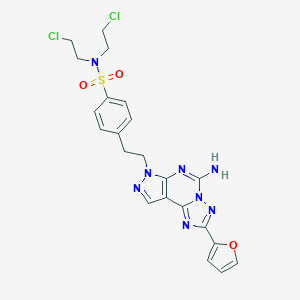 CHEMBL278805 CHEMBL278805 | C22H22Cl2N8O3S | 549.431 | 9 / 1 | 2.7 | No |
| 4238 | 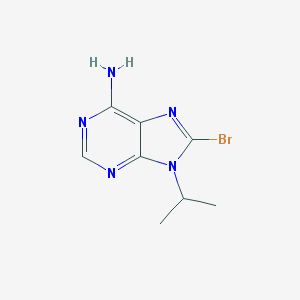 CHEMBL446264 CHEMBL446264 | C8H10BrN5 | 256.107 | 4 / 1 | 1.4 | Yes |
| 4332 | 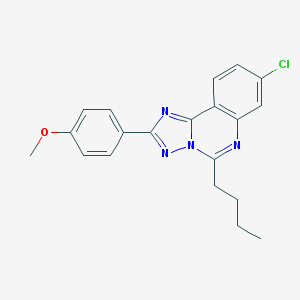 CHEMBL123669 CHEMBL123669 | C20H19ClN4O | 366.849 | 4 / 0 | 5.4 | No |
| 4353 | 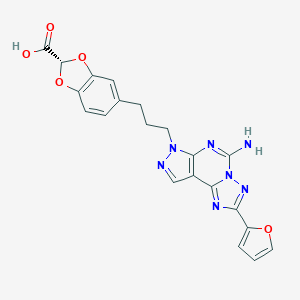 CHEMBL574701 CHEMBL574701 | C21H17N7O5 | 447.411 | 10 / 2 | 2.2 | Yes |
| 4357 | 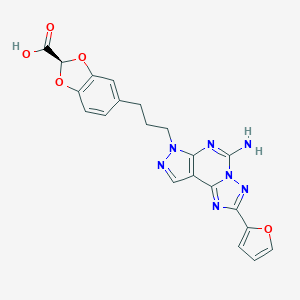 CHEMBL22355 CHEMBL22355 | C21H17N7O5 | 447.411 | 10 / 2 | 2.2 | Yes |
| 4370 | 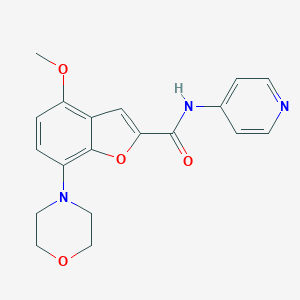 CHEMBL1082391 CHEMBL1082391 | C19H19N3O4 | 353.378 | 6 / 1 | 2.1 | Yes |
| 4384 | 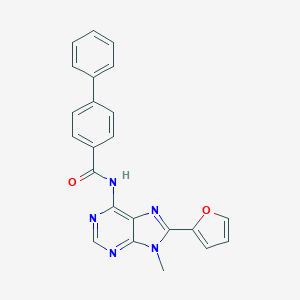 CHEMBL3087709 CHEMBL3087709 | C23H17N5O2 | 395.422 | 5 / 1 | 3.6 | Yes |
| 4438 | 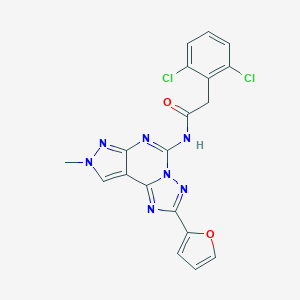 CHEMBL574490 CHEMBL574490 | C19H13Cl2N7O2 | 442.26 | 6 / 1 | 3.2 | Yes |
| 4512 | 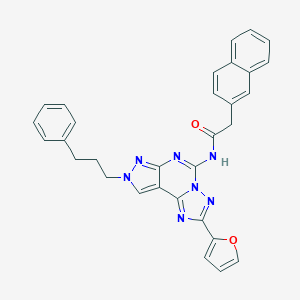 CHEMBL575605 CHEMBL575605 | C31H25N7O2 | 527.588 | 6 / 1 | 5.4 | No |
| 441888 | 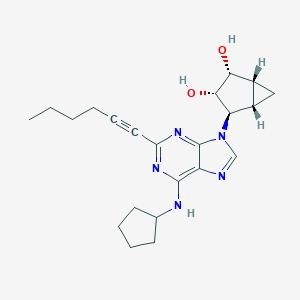 CHEMBL3125715 CHEMBL3125715 | C22H29N5O2 | 395.507 | 6 / 3 | 3.1 | Yes |
| 521592 | 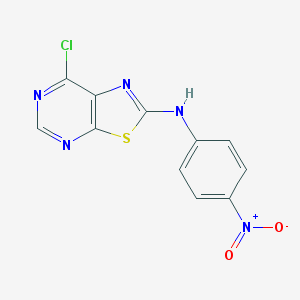 871266-83-0 871266-83-0 | C11H6ClN5O2S | 307.712 | 7 / 1 | 3.7 | Yes |
| 4753 | 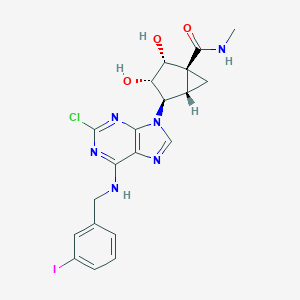 CHEMBL27376 CHEMBL27376 | C20H20ClIN6O3 | 554.773 | 7 / 4 | 2.0 | No |
| 4758 | 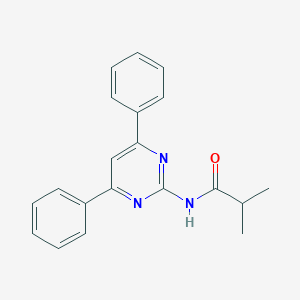 CHEMBL376778 CHEMBL376778 | C20H19N3O | 317.392 | 3 / 1 | 4.1 | Yes |
| 4817 | 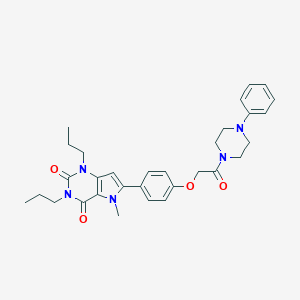 CHEMBL1171381 CHEMBL1171381 | C31H37N5O4 | 543.668 | 5 / 0 | 4.4 | No |
| 4867 | 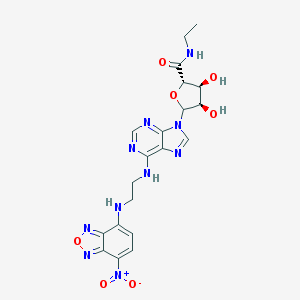 CHEMBL611552 CHEMBL611552 | C20H22N10O7 | 514.459 | 14 / 5 | -0.1 | No |
| 4868 | 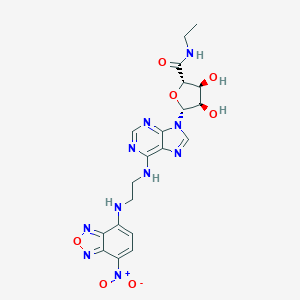 CHEMBL2413111 CHEMBL2413111 | C20H22N10O7 | 514.459 | 14 / 5 | -0.1 | No |
| 4870 | 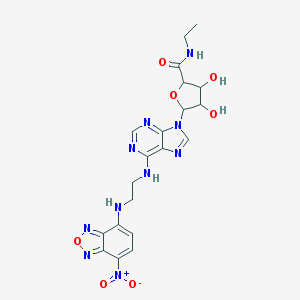 CHEMBL611552 CHEMBL611552 | C20H22N10O7 | 514.459 | 14 / 5 | -0.1 | No |
| 4878 | 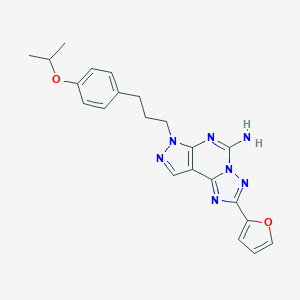 CHEMBL1258192 CHEMBL1258192 | C22H23N7O2 | 417.473 | 7 / 1 | 3.5 | Yes |
| 4934 | 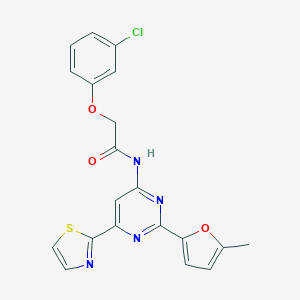 CHEMBL255861 CHEMBL255861 | C20H15ClN4O3S | 426.875 | 7 / 1 | 3.9 | Yes |
| 4967 | 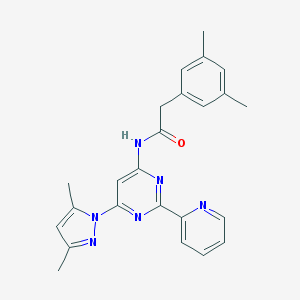 CHEMBL429420 CHEMBL429420 | C24H24N6O | 412.497 | 5 / 1 | 3.8 | Yes |
| 4996 | 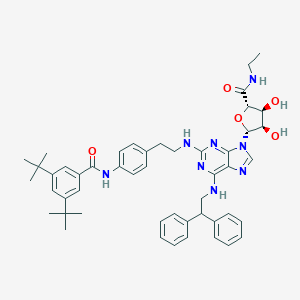 CHEMBL2042308 CHEMBL2042308 | C49H58N8O5 | 839.054 | 10 / 6 | 9.0 | No |
| 4997 | 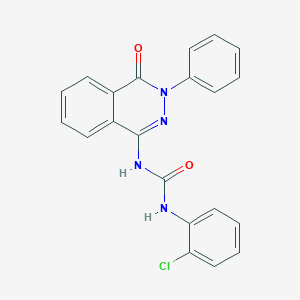 CHEMBL1766351 CHEMBL1766351 | C21H15ClN4O2 | 390.827 | 3 / 2 | 4.1 | Yes |
| 5152 | 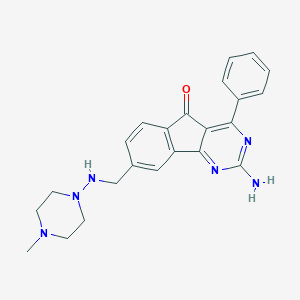 CHEMBL1088665 CHEMBL1088665 | C23H24N6O | 400.486 | 7 / 2 | 2.1 | Yes |
| 5194 | 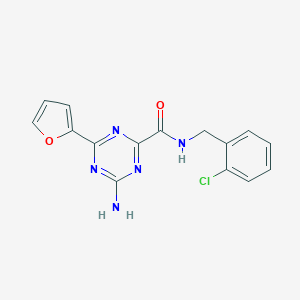 triazine-4-carboxamide, 6 triazine-4-carboxamide, 6 | C15H12ClN5O2 | 329.744 | 6 / 2 | 2.1 | Yes |
| 441910 | 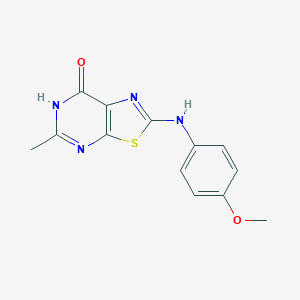 CHEMBL3415043 CHEMBL3415043 | C13H12N4O2S | 288.325 | 6 / 2 | 2.2 | Yes |
| 463519 | 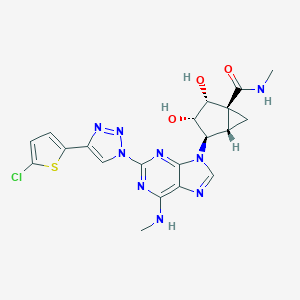 CHEMBL3612943 CHEMBL3612943 | C20H20ClN9O3S | 501.95 | 10 / 4 | 1.0 | No |
| 5372 | 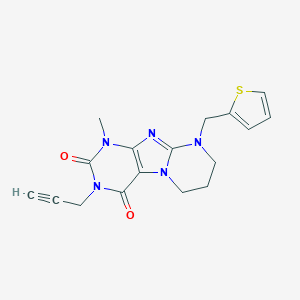 CHEMBL3093315 CHEMBL3093315 | C17H17N5O2S | 355.416 | 5 / 0 | 1.4 | Yes |
| 5402 | 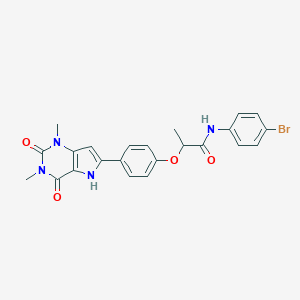 CHEMBL370588 CHEMBL370588 | C23H21BrN4O4 | 497.349 | 4 / 2 | 3.6 | Yes |
| 5608 | 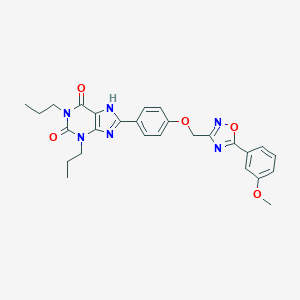 UNII-29S2U9DRGS UNII-29S2U9DRGS | C27H28N6O5 | 516.558 | 8 / 1 | 5.7 | No |
| 5624 | 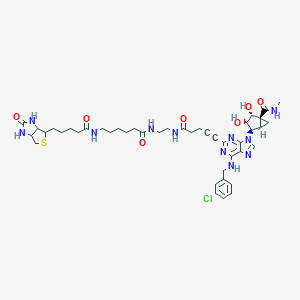 CHEMBL589513 CHEMBL589513 | C43H56ClN11O7S | 906.501 | 12 / 9 | 1.2 | No |
| 5626 | 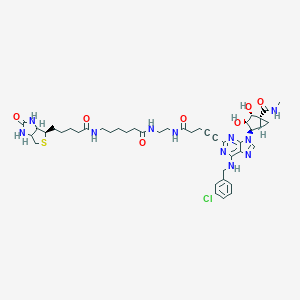 CHEMBL583748 CHEMBL583748 | C43H56ClN11O7S | 906.501 | 12 / 9 | 1.2 | No |
| 5630 | 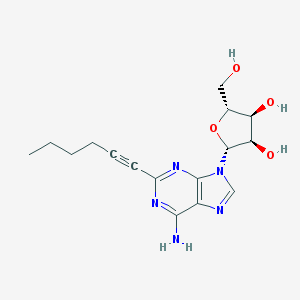 2-(1-hexynyl)adenosine 2-(1-hexynyl)adenosine | C16H21N5O4 | 347.375 | 8 / 4 | 1.3 | Yes |
| 547959 | 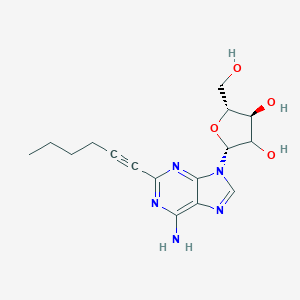 CHEMBL3981215 CHEMBL3981215 | C16H21N5O4 | 347.375 | 8 / 4 | 1.3 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417