

You can:
| Name | Melanocortin receptor 5 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | MC5R |
| Synonym | melanocortin receptor 5 MC5R MC5-R MC5 receptor MC-2 |
| Disease | Acne vulgaris Seborrhea |
| Length | 325 |
| Amino acid sequence | MNSSFHLHFLDLNLNATEGNLSGPNVKNKSSPCEDMGIAVEVFLTLGVISLLENILVIGAIVKNKNLHSPMYFFVCSLAVADMLVSMSSAWETITIYLLNNKHLVIADAFVRHIDNVFDSMICISVVASMCSLLAIAVDRYVTIFYALRYHHIMTARRSGAIIAGIWAFCTGCGIVFILYSESTYVILCLISMFFAMLFLLVSLYIHMFLLARTHVKRIAALPGASSARQRTSMQGAVTVTMLLGVFTVCWAPFFLHLTLMLSCPQNLYCSRFMSHFNMYLILIMCNSVMDPLIYAFRSQEMRKTFKEIICCRGFRIACSFPRRD |
| UniProt | P33032 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P33032 |
| 3D structure model | This predicted structure model is from GPCR-EXP P33032. |
| BioLiP | N/A |
| Therapeutic Target Database | T95302 |
| ChEMBL | CHEMBL4608 |
| IUPHAR | 286 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 2350 | 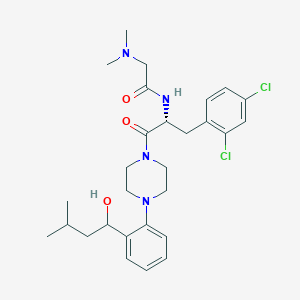 CHEMBL454916 CHEMBL454916 | C28H38Cl2N4O3 | 549.537 | 5 / 2 | 4.8 | No |
| 533920 | 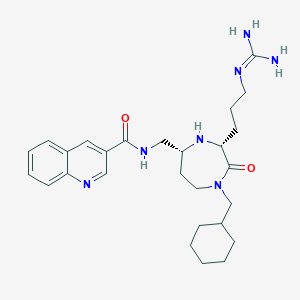 CHEMBL3949898 CHEMBL3949898 | C27H39N7O2 | 493.656 | 5 / 4 | 2.3 | Yes |
| 3808 | 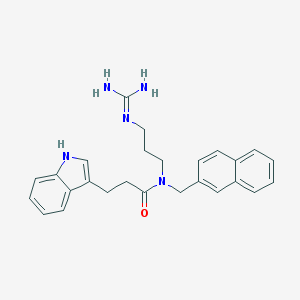 CHEMBL235798 CHEMBL235798 | C26H29N5O | 427.552 | 2 / 3 | 3.3 | Yes |
| 3967 | 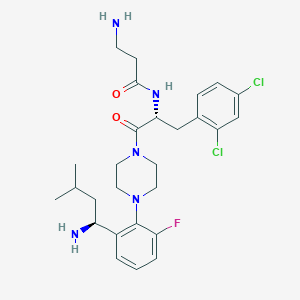 CHEMBL236014 CHEMBL236014 | C27H36Cl2FN5O2 | 552.516 | 6 / 3 | 3.6 | No |
| 441863 |  CHEMBL1775057 CHEMBL1775057 | C47H59N13O9 | 950.071 | 10 / 12 | -0.8 | No |
| 441869 |  CHEMBL1775063 CHEMBL1775063 | C47H59N13O9 | 950.071 | 10 / 12 | -0.8 | No |
| 441872 |  CHEMBL1775064 CHEMBL1775064 | C47H59N13O9 | 950.071 | 10 / 12 | -0.8 | No |
| 4668 |  120-78-5 120-78-5 | C14H8N2S4 | 332.472 | 6 / 0 | 5.6 | No |
| 4710 | 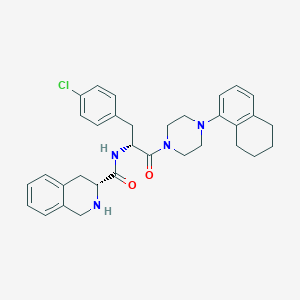 CHEMBL2113144 CHEMBL2113144 | C33H37ClN4O2 | 557.135 | 4 / 2 | 5.8 | No |
| 533927 | 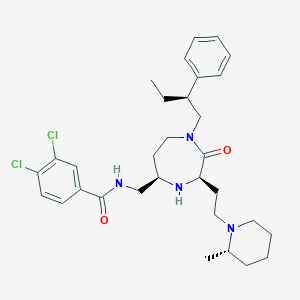 CHEMBL3978574 CHEMBL3978574 | C31H42Cl2N4O2 | 573.603 | 4 / 2 | 6.1 | No |
| 533930 | 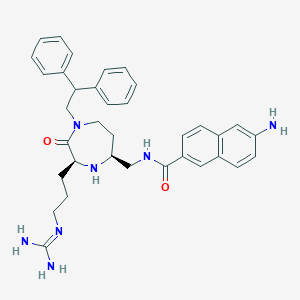 SCHEMBL2344848 SCHEMBL2344848 | C35H41N7O2 | 591.76 | 5 / 5 | 3.7 | No |
| 6468 | 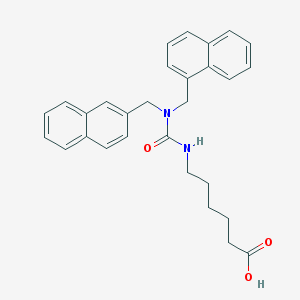 CHEMBL8825 CHEMBL8825 | C29H30N2O3 | 454.57 | 3 / 2 | 5.6 | No |
| 6523 | 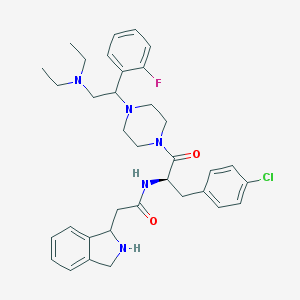 CHEMBL380855 CHEMBL380855 | C35H43ClFN5O2 | 620.21 | 6 / 2 | 4.5 | No |
| 6675 | 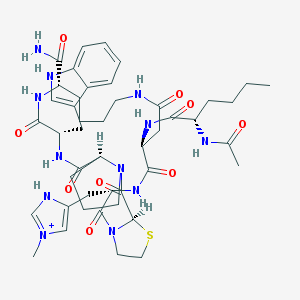 CHEMBL267360 CHEMBL267360 | C45H63N12O9S+ | 948.134 | 10 / 9 | 0.4 | No |
| 533934 | 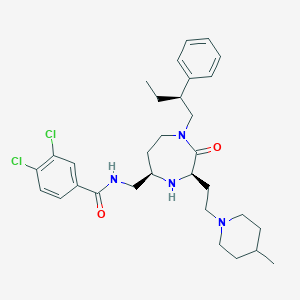 CHEMBL3944504 CHEMBL3944504 | C31H42Cl2N4O2 | 573.603 | 4 / 2 | 6.1 | No |
| 6896 | 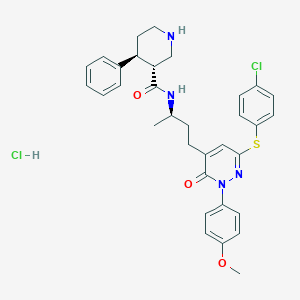 CHEMBL539306 CHEMBL539306 | C33H36Cl2N4O3S | 639.636 | 6 / 3 | N/A | No |
| 7139 | 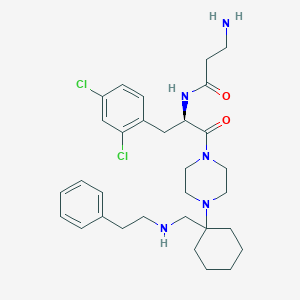 CHEMBL365237 CHEMBL365237 | C31H43Cl2N5O2 | 588.618 | 5 / 3 | 4.3 | No |
| 7589 | 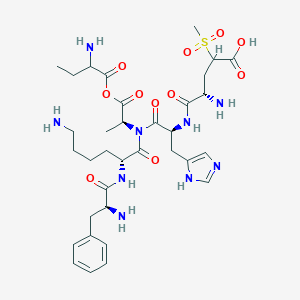 BDBM82410 BDBM82410 | C34H51N9O11S | 793.894 | 16 / 8 | -4.1 | No |
| 7706 | 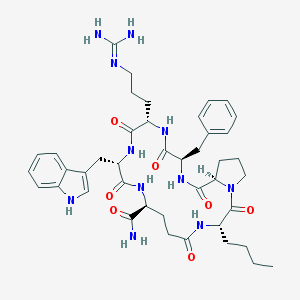 CHEMBL381739 CHEMBL381739 | C42H57N11O7 | 827.988 | 8 / 9 | 1.2 | No |
| 533940 | 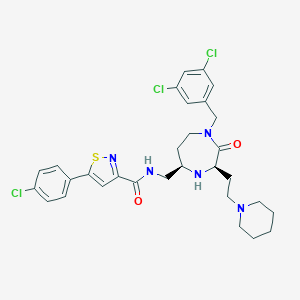 CHEMBL3918007 CHEMBL3918007 | C30H34Cl3N5O2S | 635.045 | 6 / 2 | 6.2 | No |
| 533943 | 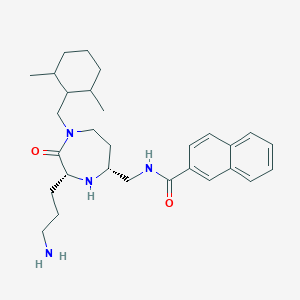 CHEMBL3931459 CHEMBL3931459 | C29H42N4O2 | 478.681 | 4 / 3 | 4.8 | Yes |
| 8177 | 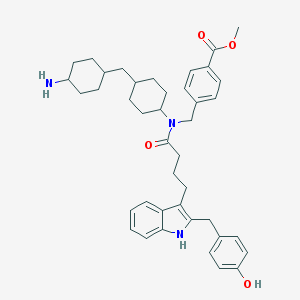 CHEMBL445669 CHEMBL445669 | C41H51N3O4 | 649.876 | 5 / 3 | 7.7 | No |
| 8760 | 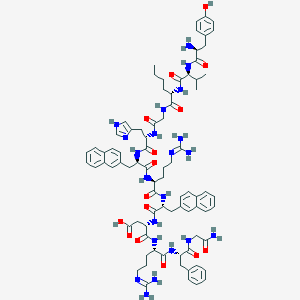 CHEMBL2370967 CHEMBL2370967 | C81H105N21O15 | 1612.86 | 19 / 20 | 0.2 | No |
| 8992 | 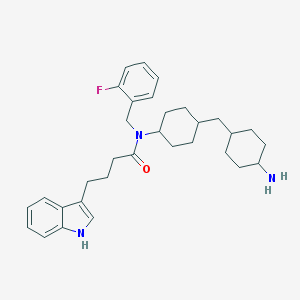 CHEMBL237070 CHEMBL237070 | C32H42FN3O | 503.706 | 3 / 2 | 6.3 | No |
| 9022 | 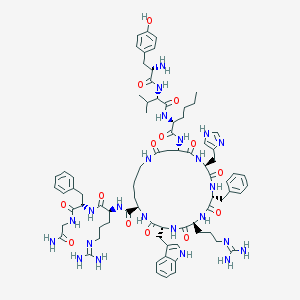 CID 44363770 CID 44363770 | C79H109N23O14 | 1604.89 | 18 / 21 | 0.7 | No |
| 533948 | 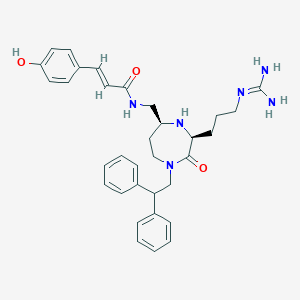 SCHEMBL2344457 SCHEMBL2344457 | C33H40N6O3 | 568.722 | 5 / 5 | 3.2 | No |
| 533949 | 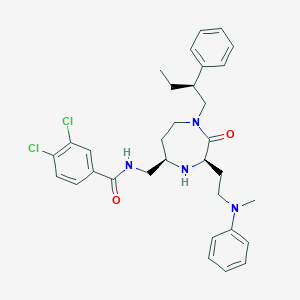 CHEMBL3973070 CHEMBL3973070 | C32H38Cl2N4O2 | 581.582 | 4 / 2 | 6.6 | No |
| 533951 | 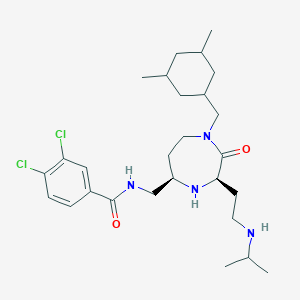 CHEMBL3964629 CHEMBL3964629 | C27H42Cl2N4O2 | 525.559 | 4 / 3 | 5.4 | No |
| 9538 |  CHEMBL1923663 CHEMBL1923663 | C44H54N12O6 | 846.994 | 8 / 10 | 1.7 | No |
| 442084 |  CHEMBL248700 CHEMBL248700 | C27H31ClN4O2 | 479.021 | 4 / 2 | 3.7 | Yes |
| 9774 |  CHEMBL237930 CHEMBL237930 | C29H31N3O | 437.587 | 2 / 2 | 4.7 | Yes |
| 533952 |  CHEMBL3931475 CHEMBL3931475 | C34H37BrN4O2 | 613.6 | 4 / 3 | 5.7 | No |
| 533953 | 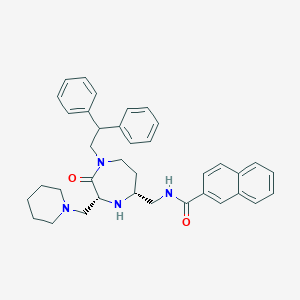 CHEMBL3979990 CHEMBL3979990 | C37H42N4O2 | 574.769 | 4 / 2 | 6.1 | No |
| 533954 | 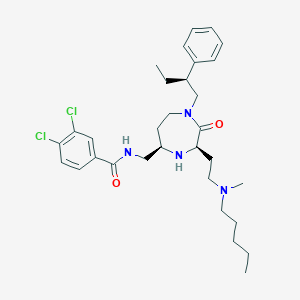 CHEMBL3948861 CHEMBL3948861 | C31H44Cl2N4O2 | 575.619 | 4 / 2 | 6.6 | No |
| 442100 | 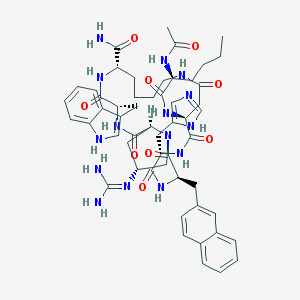 CHEMBL508501 CHEMBL508501 | C54H69N15O9 | 1072.24 | 11 / 12 | 0.9 | No |
| 10145 | 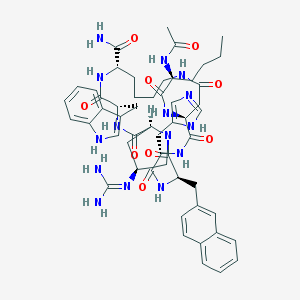 CHEMBL509582 CHEMBL509582 | C54H69N15O9 | 1072.24 | 11 / 12 | 0.9 | No |
| 10375 |  CID 16151595 CID 16151595 | C77H103N21O15 | 1562.8 | 19 / 22 | -0.3 | No |
| 533958 |  CHEMBL3912035 CHEMBL3912035 | C33H41ClN4O3 | 577.166 | 4 / 3 | 5.8 | No |
| 459325 |  CHEMBL1196971 CHEMBL1196971 | C31H48FN5O3 | 557.755 | 6 / 3 | 3.9 | No |
| 12393 |  CHEMBL365401 CHEMBL365401 | C34H51FN4O3 | 582.805 | 5 / 2 | 5.4 | No |
| 12398 | 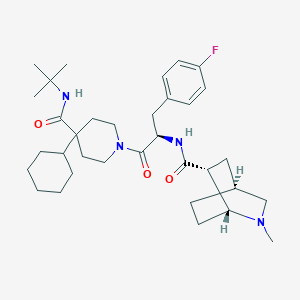 RY-764 RY-764 | C34H51FN4O3 | 582.805 | 5 / 2 | 5.4 | No |
| 12404 | 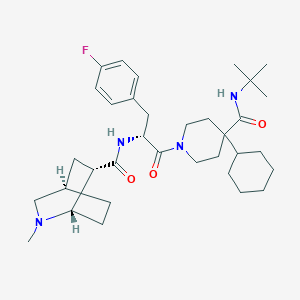 CHEMBL429387 CHEMBL429387 | C34H51FN4O3 | 582.805 | 5 / 2 | 5.4 | No |
| 12892 | 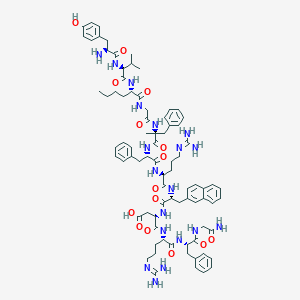 CHEMBL2370965 CHEMBL2370965 | C81H107N19O15 | 1586.86 | 18 / 19 | 0.9 | No |
| 12913 | 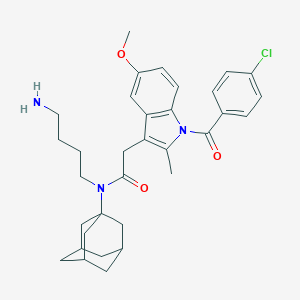 CHEMBL392094 CHEMBL392094 | C33H40ClN3O3 | 562.151 | 4 / 1 | 6.7 | No |
| 533968 | 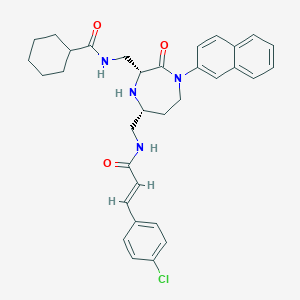 CHEMBL3958801 CHEMBL3958801 | C33H37ClN4O3 | 573.134 | 4 / 3 | 6.3 | No |
| 533970 | 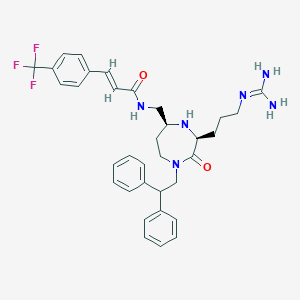 SCHEMBL2348895 SCHEMBL2348895 | C34H39F3N6O2 | 620.721 | 7 / 4 | 4.4 | No |
| 13407 | 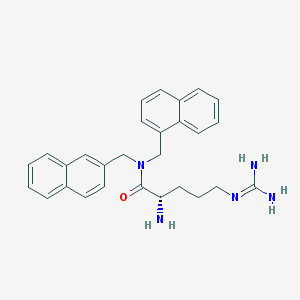 CHEMBL267020 CHEMBL267020 | C28H31N5O | 453.59 | 3 / 3 | 3.5 | Yes |
| 13525 | 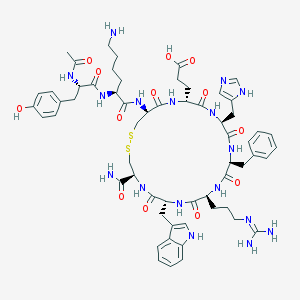 Ac-YK[CEHdFRWC]-NH2 Ac-YK[CEHdFRWC]-NH2 | C60H79N17O13S2 | 1310.52 | 18 / 17 | -3.3 | No |
| 14700 | 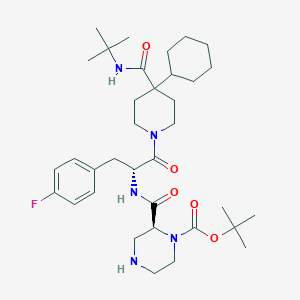 CHEMBL2113028 CHEMBL2113028 | C35H54FN5O5 | 643.845 | 7 / 3 | 4.8 | No |
| 442214 | 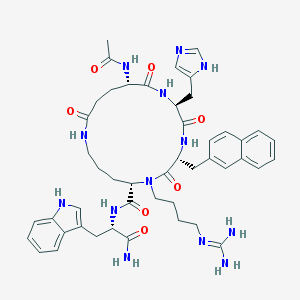 CHEMBL385561 CHEMBL385561 | C47H59N13O7 | 918.073 | 9 / 10 | 0.9 | No |
| 15526 | 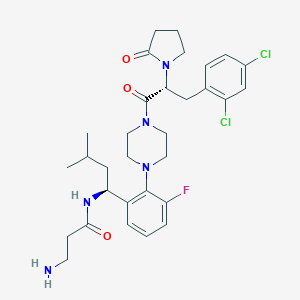 CHEMBL397580 CHEMBL397580 | C31H40Cl2FN5O3 | 620.591 | 6 / 2 | 4.1 | No |
| 15743 | 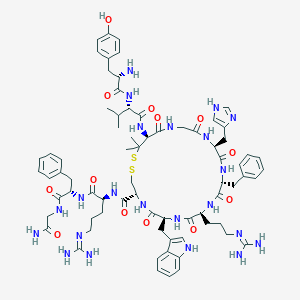 CID 44363975 CID 44363975 | C73H98N22O13S2 | 1555.85 | 19 / 20 | -0.2 | No |
| 15895 | 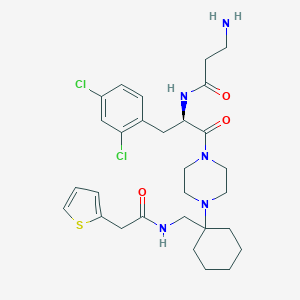 CHEMBL188651 CHEMBL188651 | C29H39Cl2N5O3S | 608.623 | 6 / 3 | 3.4 | No |
| 464869 | 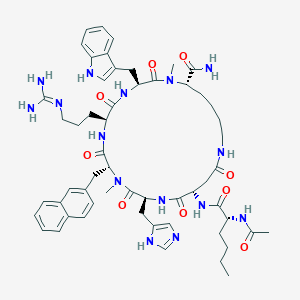 CHEMBL3600838 CHEMBL3600838 | C56H75N15O9 | 1102.31 | 11 / 11 | 1.6 | No |
| 16214 | 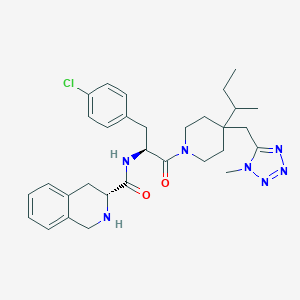 CHEMBL400456 CHEMBL400456 | C31H40ClN7O2 | 578.158 | 6 / 2 | 5.0 | No |
| 16537 | 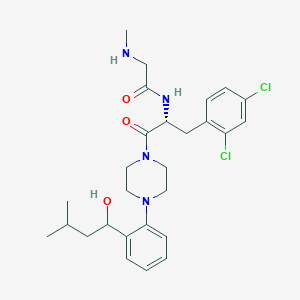 CHEMBL471340 CHEMBL471340 | C27H36Cl2N4O3 | 535.51 | 5 / 3 | 4.3 | No |
| 533975 | 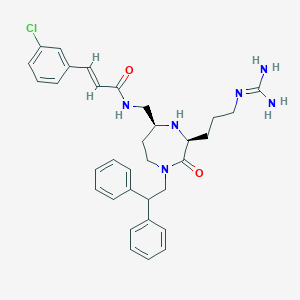 SCHEMBL2348398 SCHEMBL2348398 | C33H39ClN6O2 | 587.165 | 4 / 4 | 4.2 | No |
| 17527 | 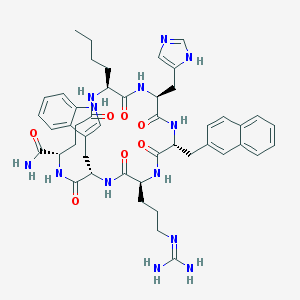 CHEMBL425591 CHEMBL425591 | C47H59N13O7 | 918.073 | 9 / 11 | 1.9 | No |
| 533978 | 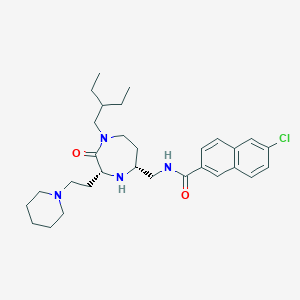 CHEMBL3890817 CHEMBL3890817 | C30H43ClN4O2 | 527.15 | 4 / 2 | 5.7 | No |
| 18246 | 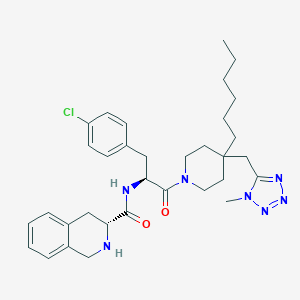 CHEMBL249277 CHEMBL249277 | C33H44ClN7O2 | 606.212 | 6 / 2 | 6.3 | No |
| 18607 | 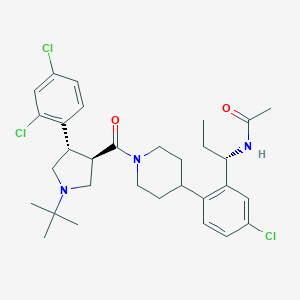 CHEMBL254757 CHEMBL254757 | C31H40Cl3N3O2 | 593.03 | 3 / 1 | 6.4 | No |
| 18756 | 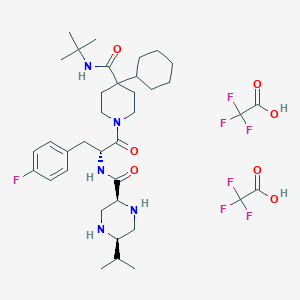 CHEMBL2028957 CHEMBL2028957 | C37H54F7N5O7 | 813.856 | 16 / 6 | N/A | No |
| 465272 | 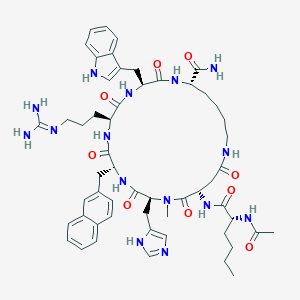 CHEMBL3600835 CHEMBL3600835 | C55H73N15O9 | 1088.29 | 11 / 12 | 1.5 | No |
| 19968 | 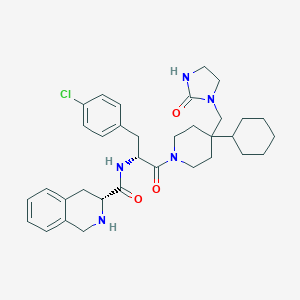 CHEMBL381503 CHEMBL381503 | C34H44ClN5O3 | 606.208 | 4 / 3 | 5.3 | No |
| 20122 | 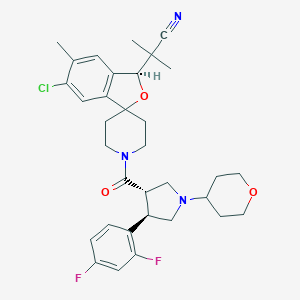 CHEMBL1209788 CHEMBL1209788 | C33H38ClF2N3O3 | 598.132 | 7 / 0 | 4.8 | No |
| 533990 | 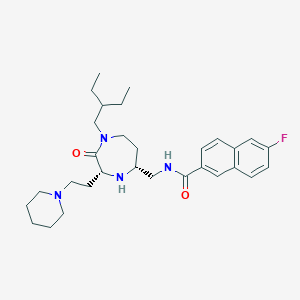 CHEMBL3906338 CHEMBL3906338 | C30H43FN4O2 | 510.698 | 5 / 2 | 5.2 | No |
| 21529 | 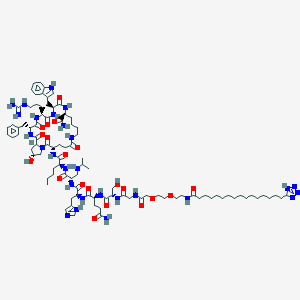 CID 70693082 CID 70693082 | C93H145N27O20 | 1961.35 | 26 / 24 | 1.5 | No |
| 533993 | 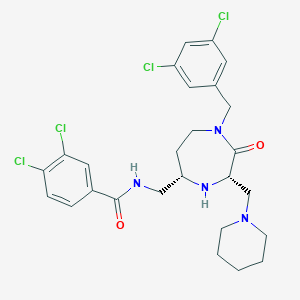 SCHEMBL2351596 SCHEMBL2351596 | C26H30Cl4N4O2 | 572.352 | 4 / 2 | 5.4 | No |
| 533995 | 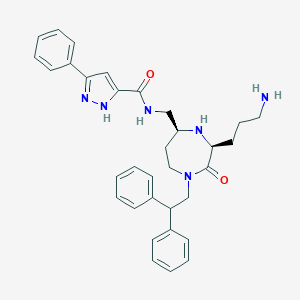 SCHEMBL2349417 SCHEMBL2349417 | C33H38N6O2 | 550.707 | 5 / 4 | 4.0 | No |
| 22162 | 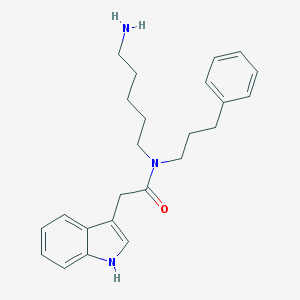 CHEMBL237052 CHEMBL237052 | C24H31N3O | 377.532 | 2 / 2 | 3.8 | Yes |
| 533999 | 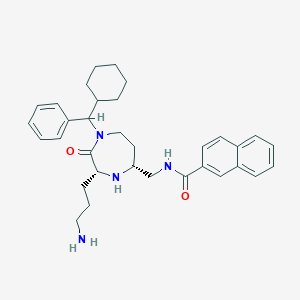 CHEMBL3958082 CHEMBL3958082 | C33H42N4O2 | 526.725 | 4 / 3 | 5.5 | No |
| 23189 | 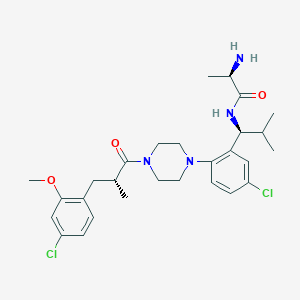 CHEMBL467587 CHEMBL467587 | C28H38Cl2N4O3 | 549.537 | 5 / 2 | 5.0 | No |
| 23309 | 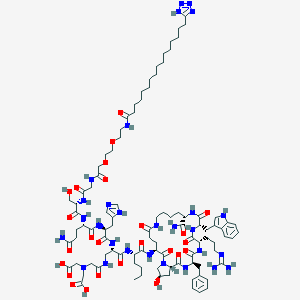 BDBM50389782 BDBM50389782 | C96H146N28O25 | 2092.4 | 31 / 25 | -3.0 | No |
| 534002 | 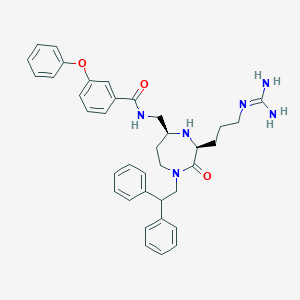 SCHEMBL2347568 SCHEMBL2347568 | C37H42N6O3 | 618.782 | 5 / 4 | 4.6 | No |
| 23783 | 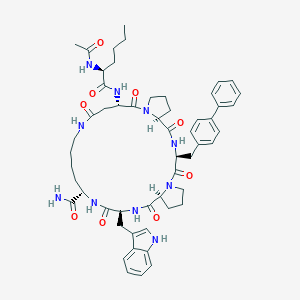 CHEMBL2371963 CHEMBL2371963 | C54H68N10O9 | 1001.2 | 9 / 8 | 3.7 | No |
| 534004 | 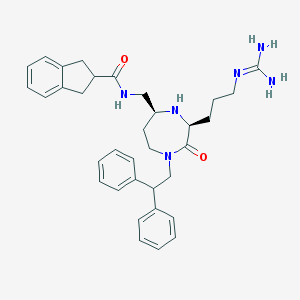 SCHEMBL2347332 SCHEMBL2347332 | C34H42N6O2 | 566.75 | 4 / 4 | 3.4 | No |
| 534005 | 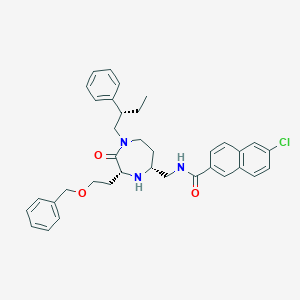 CHEMBL3962578 CHEMBL3962578 | C36H40ClN3O3 | 598.184 | 4 / 2 | 6.8 | No |
| 24583 | 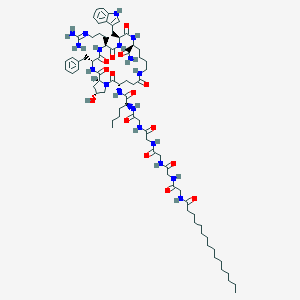 CHEMBL2070248 CHEMBL2070248 | C74H114N18O15 | 1495.84 | 16 / 17 | 4.0 | No |
| 24643 | 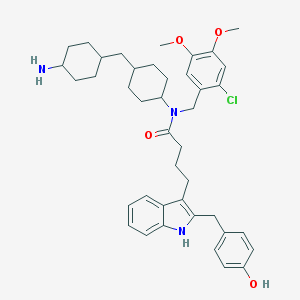 CHEMBL396111 CHEMBL396111 | C41H52ClN3O4 | 686.334 | 5 / 3 | 8.4 | No |
| 24968 | 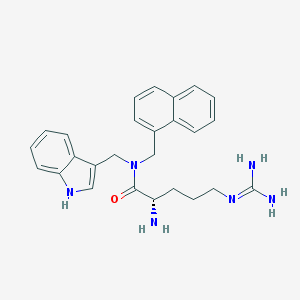 CHEMBL2113280 CHEMBL2113280 | C26H30N6O | 442.567 | 3 / 4 | 2.4 | Yes |
| 25995 | 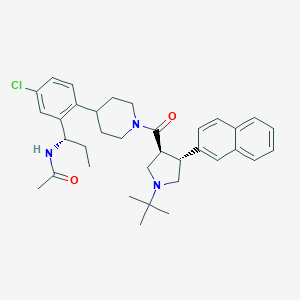 CHEMBL403420 CHEMBL403420 | C35H44ClN3O2 | 574.206 | 3 / 1 | 6.4 | No |
| 534010 | 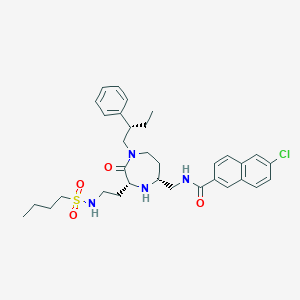 CHEMBL3975125 CHEMBL3975125 | C33H43ClN4O4S | 627.241 | 6 / 3 | 5.8 | No |
| 466223 | 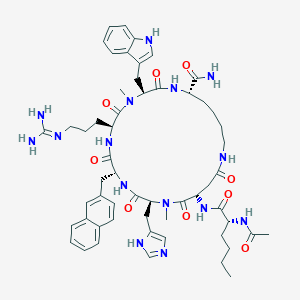 CHEMBL3600842 CHEMBL3600842 | C56H75N15O9 | 1102.31 | 11 / 11 | 1.6 | No |
| 27257 | 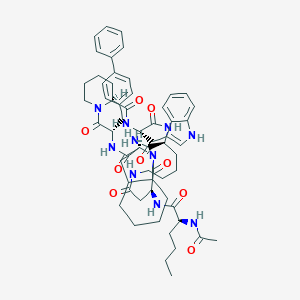 CHEMBL2371968 CHEMBL2371968 | C59H76N10O9 | 1069.32 | 9 / 8 | 5.4 | No |
| 27694 | 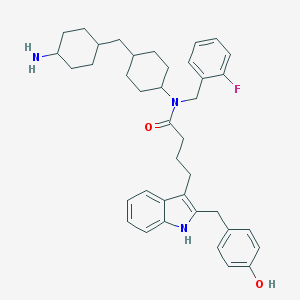 CHEMBL394394 CHEMBL394394 | C39H48FN3O2 | 609.83 | 4 / 3 | 7.9 | No |
| 28060 | 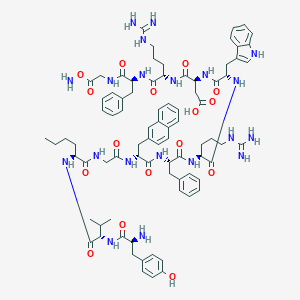 BDBM50163151 BDBM50163151 | C82H106N20O16 | 1627.87 | 20 / 22 | 1.8 | No |
| 29094 | 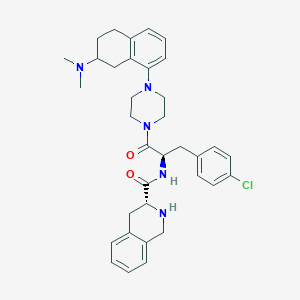 CHEMBL2113149 CHEMBL2113149 | C35H42ClN5O2 | 600.204 | 5 / 2 | 5.2 | No |
| 29378 | 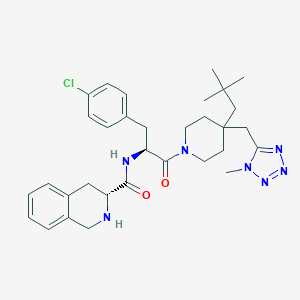 CHEMBL400252 CHEMBL400252 | C32H42ClN7O2 | 592.185 | 6 / 2 | 5.4 | No |
| 534022 | 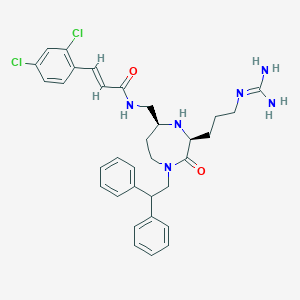 SCHEMBL2349021 SCHEMBL2349021 | C33H38Cl2N6O2 | 621.607 | 4 / 4 | 4.8 | No |
| 534032 | 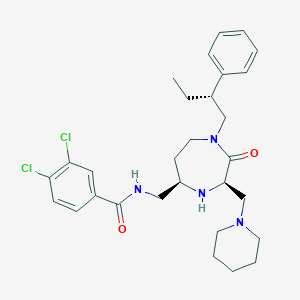 CHEMBL3916858 CHEMBL3916858 | C29H38Cl2N4O2 | 545.549 | 4 / 2 | 5.3 | No |
| 31832 | 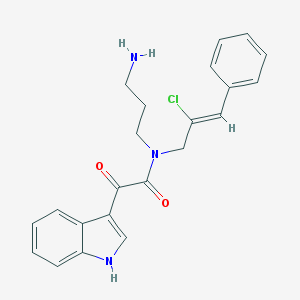 CHEMBL237937 CHEMBL237937 | C22H22ClN3O2 | 395.887 | 3 / 2 | 3.4 | Yes |
| 534037 | 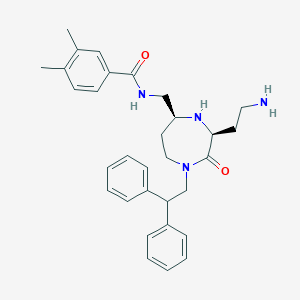 SCHEMBL2347544 SCHEMBL2347544 | C31H38N4O2 | 498.671 | 4 / 3 | 4.1 | Yes |
| 33381 | 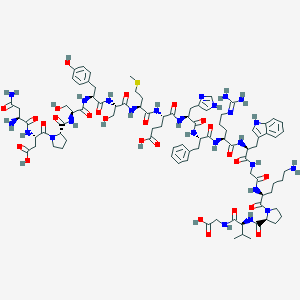 D0I8SJ D0I8SJ | C90H127N25O26S | 2007.21 | 31 / 27 | -10.1 | No |
| 33646 | 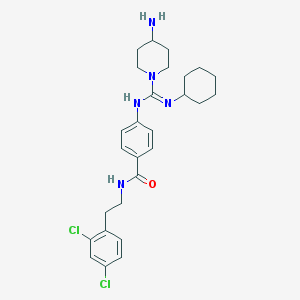 CHEMBL366036 CHEMBL366036 | C27H35Cl2N5O | 516.511 | 3 / 3 | 5.1 | No |
| 442972 | 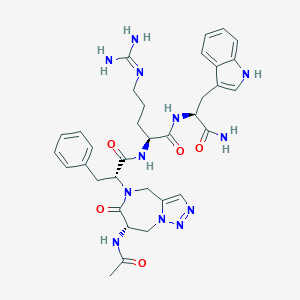 CHEMBL3421680 CHEMBL3421680 | C34H42N12O5 | 698.789 | 8 / 7 | -0.9 | No |
| 34059 | 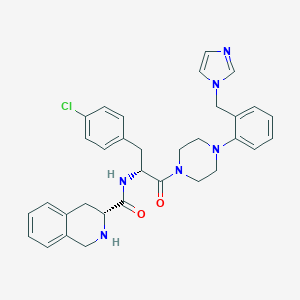 CHEMBL423101 CHEMBL423101 | C33H35ClN6O2 | 583.133 | 5 / 2 | 4.1 | No |
| 534044 | 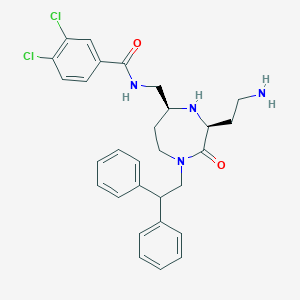 SCHEMBL2349052 SCHEMBL2349052 | C29H32Cl2N4O2 | 539.501 | 4 / 3 | 4.6 | No |
| 534046 | 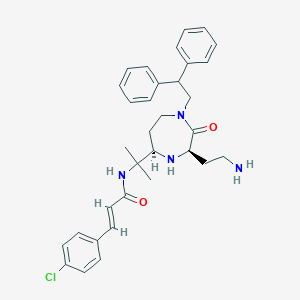 CHEMBL3954657 CHEMBL3954657 | C33H39ClN4O2 | 559.151 | 4 / 3 | 5.0 | No |
| 35071 | 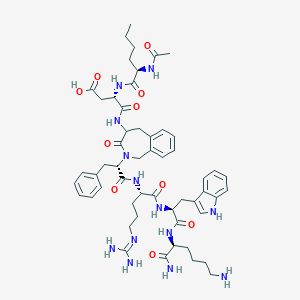 CHEMBL234456 CHEMBL234456 | C54H73N13O10 | 1064.26 | 12 / 12 | -1.1 | No |
| 35332 | 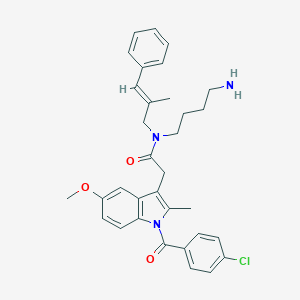 CHEMBL236840 CHEMBL236840 | C33H36ClN3O3 | 558.119 | 4 / 1 | 6.7 | No |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417