

You can:
| Name | Histamine H1 receptor |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | HRH1 |
| Synonym | HH1R H1R Hisr H1 receptor |
| Disease | Vertigo's disease; Meniere's disease Ocular allergy Obesity Nausea; Vomiting Insomnia; Anxiety disorder [ Show all ] |
| Length | 487 |
| Amino acid sequence | MSLPNSSCLLEDKMCEGNKTTMASPQLMPLVVVLSTICLVTVGLNLLVLYAVRSERKLHTVGNLYIVSLSVADLIVGAVVMPMNILYLLMSKWSLGRPLCLFWLSMDYVASTASIFSVFILCIDRYRSVQQPLRYLKYRTKTRASATILGAWFLSFLWVIPILGWNHFMQQTSVRREDKCETDFYDVTWFKVMTAIINFYLPTLLMLWFYAKIYKAVRQHCQHRELINRSLPSFSEIKLRPENPKGDAKKPGKESPWEVLKRKPKDAGGGSVLKSPSQTPKEMKSPVVFSQEDDREVDKLYCFPLDIVHMQAAAEGSSRDYVAVNRSHGQLKTDEQGLNTHGASEISEDQMLGDSQSFSRTDSDTTTETAPGKGKLRSGSNTGLDYIKFTWKRLRSHSRQYVSGLHMNRERKAAKQLGFIMAAFILCWIPYFIFFMVIAFCKNCCNEHLHMFTIWLGYINSTLNPLIYPLCNENFKKTFKRILHIRS |
| UniProt | P35367 |
| Protein Data Bank | 3rze |
| GPCR-HGmod model | P35367 |
| 3D structure model | This structure is from PDB ID 3rze. |
| BioLiP | BL0202178, BL0202179, BL0202180 |
| Therapeutic Target Database | T77913 |
| ChEMBL | CHEMBL231 |
| IUPHAR | 262 |
| DrugBank | BE0000442 |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 521461 | 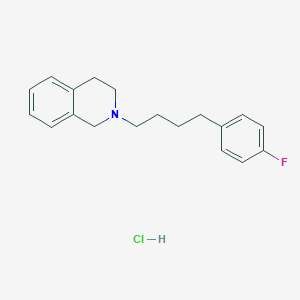 CHEMBL3818128 CHEMBL3818128 | C19H23ClFN | 319.848 | 2 / 1 | N/A | N/A |
| 765 | 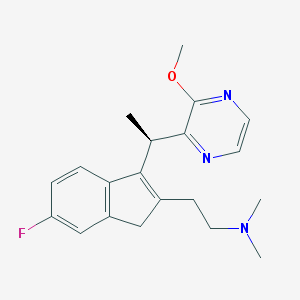 CHEMBL1090526 CHEMBL1090526 | C20H24FN3O | 341.43 | 5 / 0 | 2.7 | Yes |
| 785 | 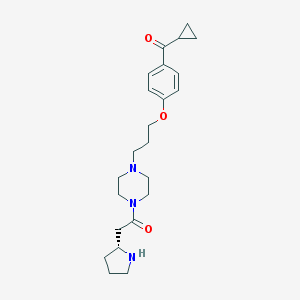 CHEMBL60435 CHEMBL60435 | C23H33N3O3 | 399.535 | 5 / 1 | 1.8 | Yes |
| 1024 | 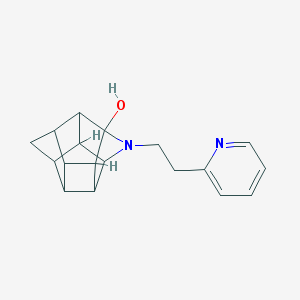 CHEMBL2205816 CHEMBL2205816 | C18H20N2O | 280.371 | 3 / 1 | 1.8 | Yes |
| 1047 | 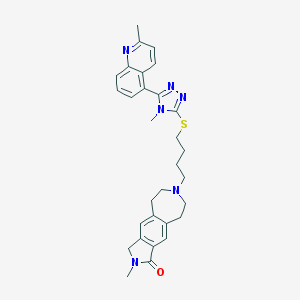 CHEMBL270853 CHEMBL270853 | C30H34N6OS | 526.703 | 6 / 0 | 4.4 | No |
| 553255 | 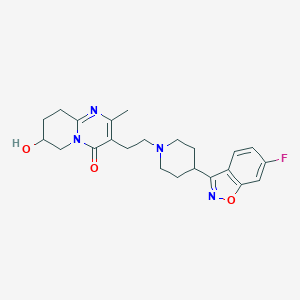 7-Hydroxy Risperidone 7-Hydroxy Risperidone | C23H27FN4O3 | 426.492 | 7 / 1 | 1.7 | Yes |
| 1256 | 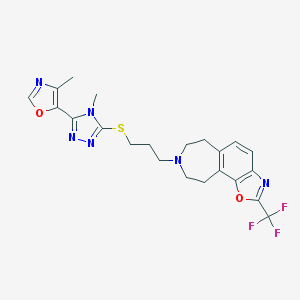 CHEMBL256659 CHEMBL256659 | C22H23F3N6O2S | 492.521 | 11 / 0 | 3.9 | No |
| 547918 | 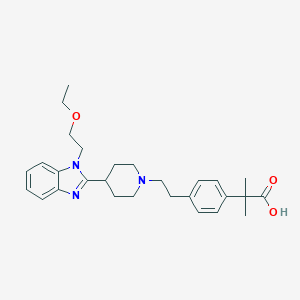 Bilastine Bilastine | C28H37N3O3 | 463.622 | 5 / 1 | 2.3 | Yes |
| 521481 | 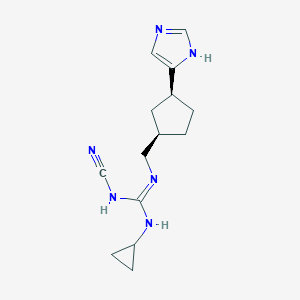 CHEMBL3806146 CHEMBL3806146 | C14H20N6 | 272.356 | 3 / 3 | 1.7 | Yes |
| 521484 | 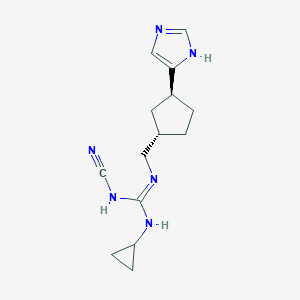 CHEMBL3804837 CHEMBL3804837 | C14H20N6 | 272.356 | 3 / 3 | 1.7 | Yes |
| 1788 | 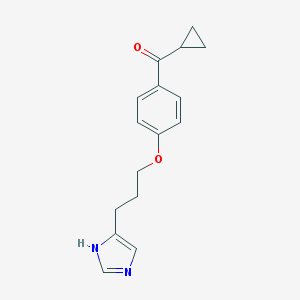 ciproxifan ciproxifan | C16H18N2O2 | 270.332 | 3 / 1 | 2.5 | Yes |
| 1840 | 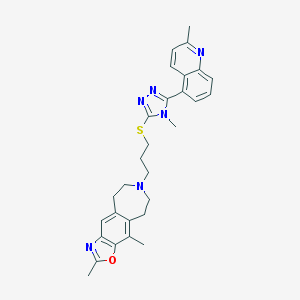 CHEMBL272694 CHEMBL272694 | C29H32N6OS | 512.676 | 7 / 0 | 5.6 | No |
| 1906 | 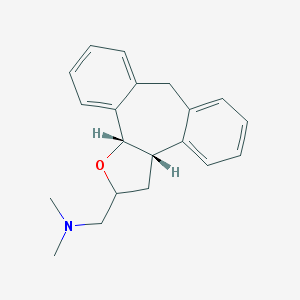 CHEMBL96732 CHEMBL96732 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1907 | 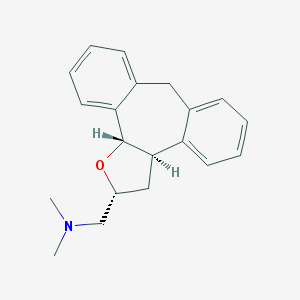 CHEMBL329566 CHEMBL329566 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1911 | 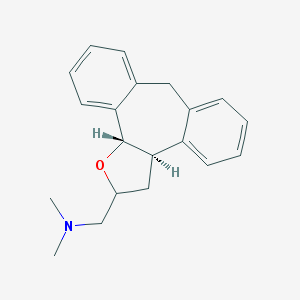 CHEMBL92667 CHEMBL92667 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1914 | 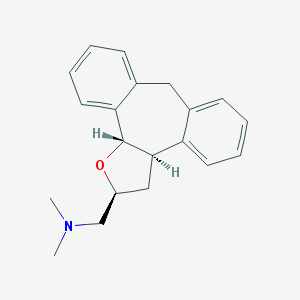 CHEMBL329268 CHEMBL329268 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1918 | 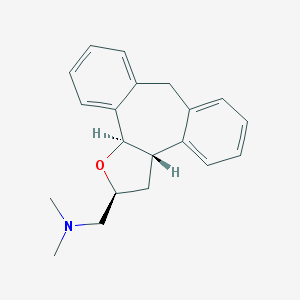 CHEMBL327651 CHEMBL327651 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 1920 | 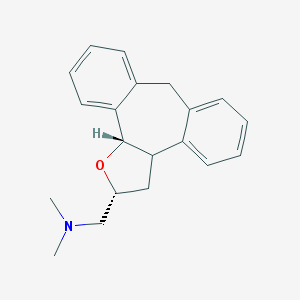 CHEMBL371352 CHEMBL371352 | C20H23NO | 293.41 | 2 / 0 | 3.6 | Yes |
| 2288 | 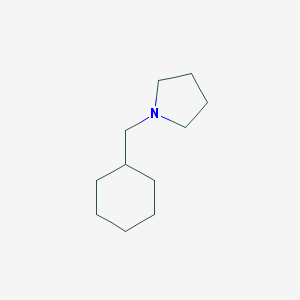 1-(cyclohexylmethyl)pyrrolidine 1-(cyclohexylmethyl)pyrrolidine | C11H21N | 167.296 | 1 / 0 | 3.1 | Yes |
| 2333 | 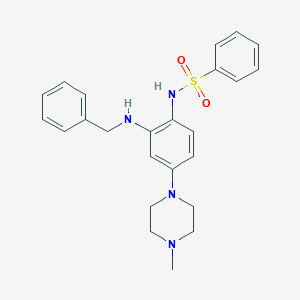 CHEMBL249797 CHEMBL249797 | C24H28N4O2S | 436.574 | 6 / 2 | 3.9 | Yes |
| 2398 | 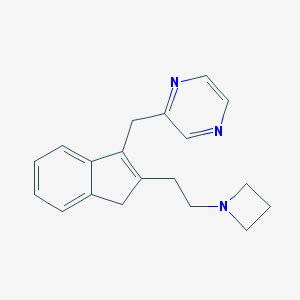 CHEMBL1669412 CHEMBL1669412 | C19H21N3 | 291.398 | 3 / 0 | 2.1 | Yes |
| 3021 | 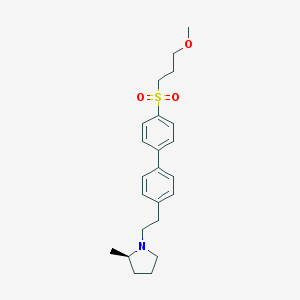 UNII-10521U3EQA UNII-10521U3EQA | C23H31NO3S | 401.565 | 4 / 0 | 4.2 | Yes |
| 521533 | 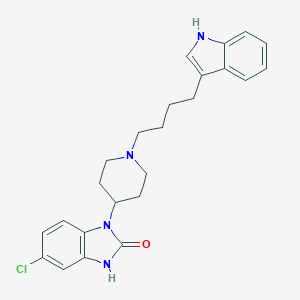 CHEMBL3818805 CHEMBL3818805 | C24H27ClN4O | 422.957 | 2 / 2 | 4.8 | Yes |
| 3145 | 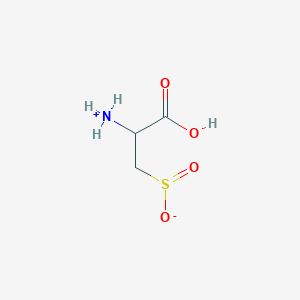 AC1N0YGA AC1N0YGA | C3H7NO4S | 153.152 | 5 / 2 | -4.7 | Yes |
| 3264 | 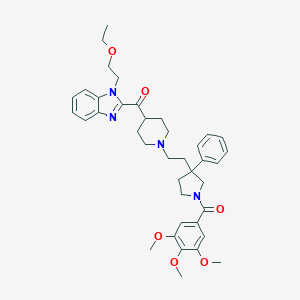 CHEMBL322194 CHEMBL322194 | C39H48N4O6 | 668.835 | 8 / 0 | 5.4 | No |
| 3326 | 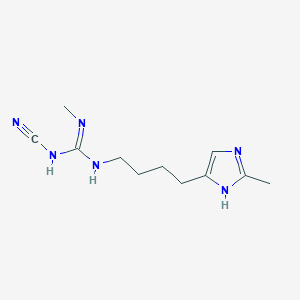 CHEMBL3220637 CHEMBL3220637 | C11H18N6 | 234.307 | 3 / 3 | 0.8 | Yes |
| 3344 | 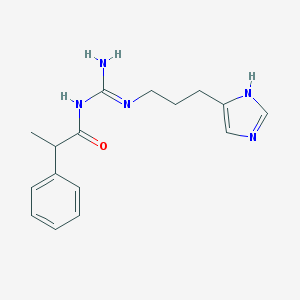 UR-AK68 UR-AK68 | C16H21N5O | 299.378 | 3 / 3 | 2.1 | Yes |
| 3697 | 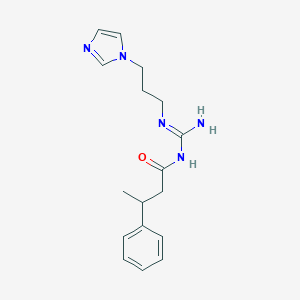 CHEMBL3220892 CHEMBL3220892 | C17H23N5O | 313.405 | 3 / 2 | 1.7 | Yes |
| 557390 | 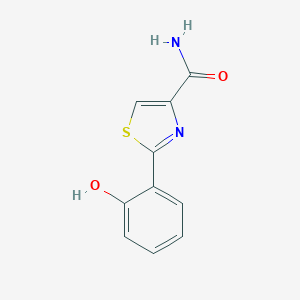 Pulicatins F Pulicatins F | C10H8N2O2S | 220.246 | 4 / 2 | 1.5 | Yes |
| 3918 | 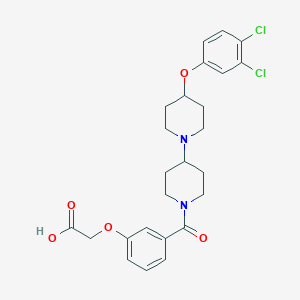 CHEMBL2171022 CHEMBL2171022 | C25H28Cl2N2O5 | 507.408 | 6 / 1 | 2.7 | No |
| 4293 | 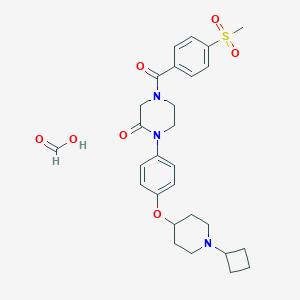 CHEMBL398996 CHEMBL398996 | C28H35N3O7S | 557.662 | 8 / 1 | N/A | No |
| 4470 | 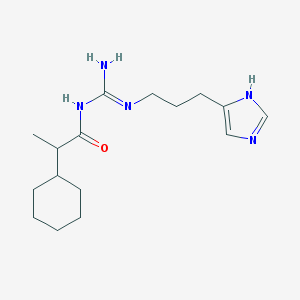 UR-AK59 UR-AK59 | C16H27N5O | 305.426 | 3 / 3 | 3.1 | Yes |
| 4890 |  CHEMBL270376 CHEMBL270376 | C23H25F3N6OS2 | 522.609 | 11 / 0 | 4.9 | No |
| 4954 |  5HT6-ligand-1 5HT6-ligand-1 | C20H22BrN3O2S | 448.379 | 4 / 0 | 3.6 | Yes |
| 5055 |  BDBM50068818 BDBM50068818 | C32H39N5O6 | 589.693 | 8 / 2 | 2.9 | No |
| 5577 |  CHEMBL330172 CHEMBL330172 | C41H45N5O5 | 687.841 | 8 / 0 | 5.8 | No |
| 5672 | 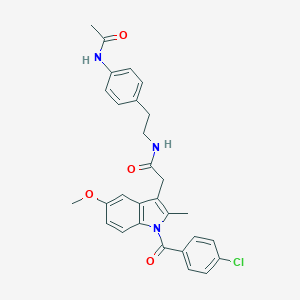 AC1L1HUK AC1L1HUK | C29H28ClN3O4 | 518.01 | 4 / 2 | 5.2 | No |
| 5751 | 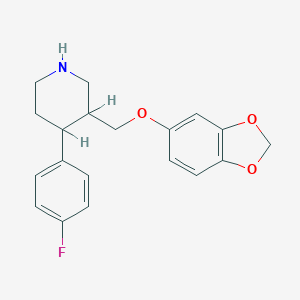 AHOUBRCZNHFOSL-UHFFFAOYSA-N AHOUBRCZNHFOSL-UHFFFAOYSA-N | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5754 | 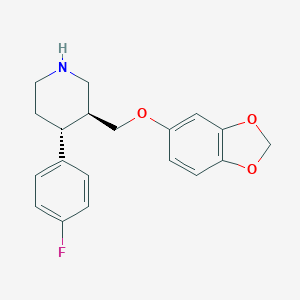 paroxetine paroxetine | C19H20FNO3 | 329.371 | 5 / 1 | 3.5 | Yes |
| 5884 | 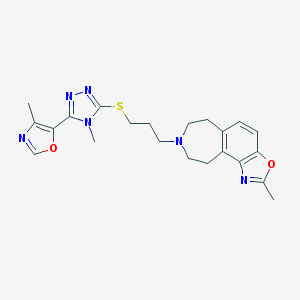 CHEMBL402422 CHEMBL402422 | C22H26N6O2S | 438.55 | 8 / 0 | 3.4 | Yes |
| 6191 | 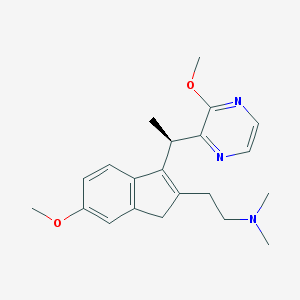 CHEMBL1090186 CHEMBL1090186 | C21H27N3O2 | 353.466 | 5 / 0 | 2.5 | Yes |
| 6458 | 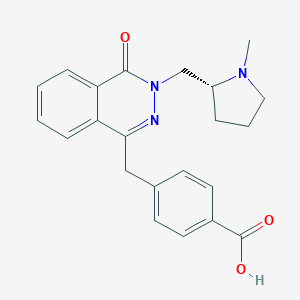 CHEMBL1767144 CHEMBL1767144 | C22H23N3O3 | 377.444 | 5 / 1 | 0.6 | Yes |
| 6507 | 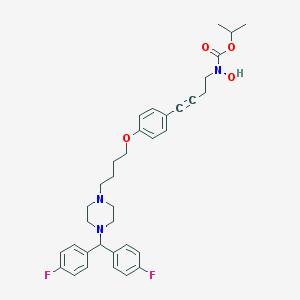 CHEMBL180233 CHEMBL180233 | C35H41F2N3O4 | 605.727 | 8 / 1 | 6.7 | No |
| 463717 | 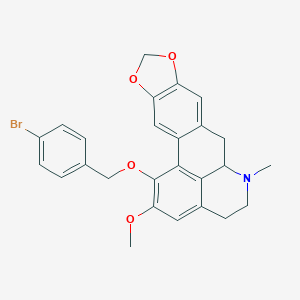 CHEMBL3633757 CHEMBL3633757 | C26H24BrNO4 | 494.385 | 5 / 0 | 5.4 | No |
| 536128 |  CHEMBL3899832 CHEMBL3899832 | C22H19N5O4 | 417.425 | 8 / 2 | 3.6 | Yes |
| 7231 |  CHEMBL1258754 CHEMBL1258754 | C19H25N3O2 | 327.428 | 3 / 1 | 2.7 | Yes |
| 7254 |  CHEMBL2208426 CHEMBL2208426 | C20H24Cl2N2O5S2 | 507.441 | 7 / 2 | 3.7 | No |
| 7430 |  CHEMBL1210209 CHEMBL1210209 | C29H35FN4O3 | 506.622 | 7 / 2 | 4.9 | No |
| 7469 |  N-Methyl-3,3-diphenylpropylamine N-Methyl-3,3-diphenylpropylamine | C16H19N | 225.335 | 1 / 1 | 3.6 | Yes |
| 7889 |  CHEMBL158352 CHEMBL158352 | C14H27N3 | 237.391 | 2 / 2 | 3.7 | Yes |
| 8021 |  CHEMBL1258755 CHEMBL1258755 | C20H27N3O2 | 341.455 | 3 / 1 | 3.0 | Yes |
| 8211 |  BW-723C86 BW-723C86 | C16H18N2OS | 286.393 | 3 / 2 | 3.0 | Yes |
| 8469 | 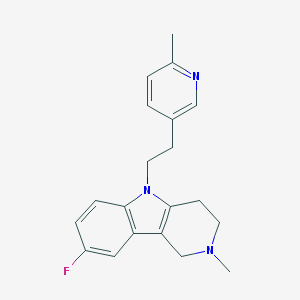 CHEMBL1783963 CHEMBL1783963 | C20H22FN3 | 323.415 | 3 / 0 | 3.2 | Yes |
| 8521 | 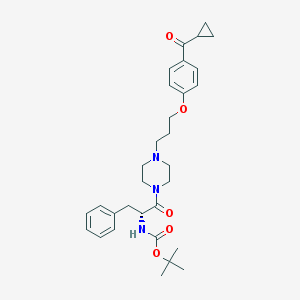 CHEMBL60143 CHEMBL60143 | C31H41N3O5 | 535.685 | 6 / 1 | 4.4 | No |
| 521687 | 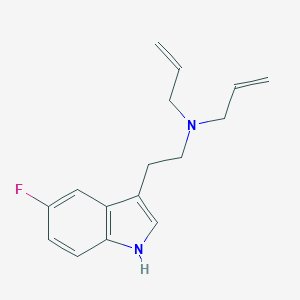 CHEMBL3754496 CHEMBL3754496 | C16H19FN2 | 258.34 | 2 / 1 | 2.9 | Yes |
| 8615 | 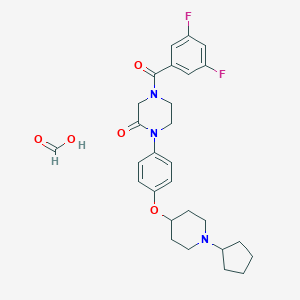 CHEMBL240567 CHEMBL240567 | C28H33F2N3O5 | 529.585 | 8 / 1 | N/A | No |
| 442053 | 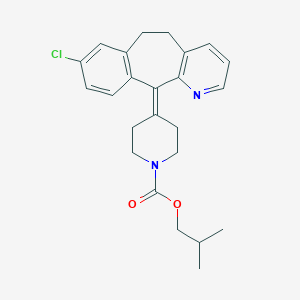 CHEMBL3357035 CHEMBL3357035 | C24H27ClN2O2 | 410.942 | 3 / 0 | 6.2 | No |
| 8811 | 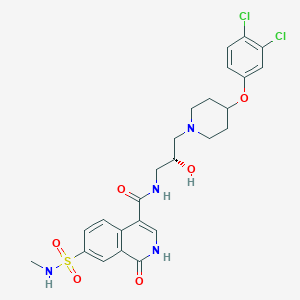 CHEMBL2207284 CHEMBL2207284 | C25H28Cl2N4O6S | 583.481 | 8 / 4 | 2.2 | No |
| 9161 | 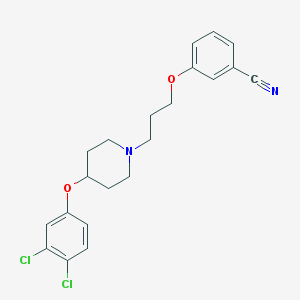 CHEMBL2208420 CHEMBL2208420 | C21H22Cl2N2O2 | 405.319 | 4 / 0 | 5.5 | No |
| 9853 | 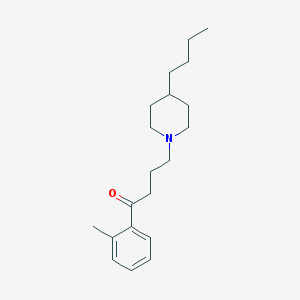 AC-42 AC-42 | C20H31NO | 301.474 | 2 / 0 | 5.2 | No |
| 557568 | 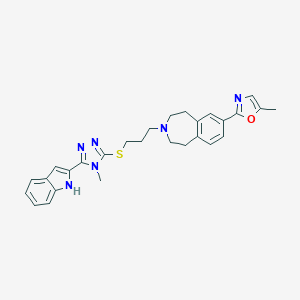 CHEMBL397429 CHEMBL397429 | C28H30N6OS | 498.649 | 6 / 1 | 5.0 | Yes |
| 10180 | 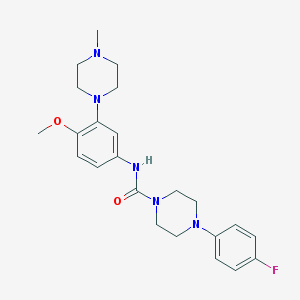 CHEMBL194206 CHEMBL194206 | C23H30FN5O2 | 427.524 | 6 / 1 | 2.7 | Yes |
| 10381 | 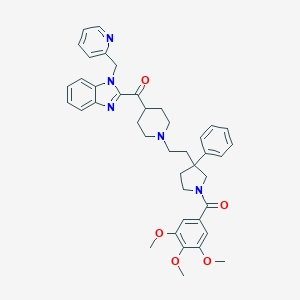 CHEMBL96442 CHEMBL96442 | C41H45N5O5 | 687.841 | 8 / 0 | 5.8 | No |
| 10564 | 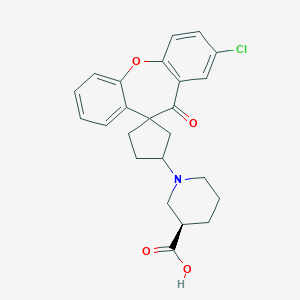 CHEMBL1277492 CHEMBL1277492 | C24H24ClNO4 | 425.909 | 5 / 1 | 2.1 | Yes |
| 10744 |  CHEMBL3287723 CHEMBL3287723 | C25H32O7 | 444.524 | 7 / 2 | 5.7 | No |
| 11248 |  CHEMBL494982 CHEMBL494982 | C17H21N3O | 283.375 | 3 / 1 | 2.9 | Yes |
| 11512 |  CHEMBL1257376 CHEMBL1257376 | C26H30FN3O2 | 435.543 | 4 / 0 | 4.5 | Yes |
| 11739 |  propranolol propranolol | C16H21NO2 | 259.349 | 3 / 2 | 3.0 | Yes |
| 11791 | 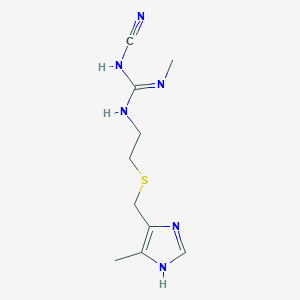 cimetidine cimetidine | C10H16N6S | 252.34 | 4 / 3 | 0.4 | Yes |
| 11802 | 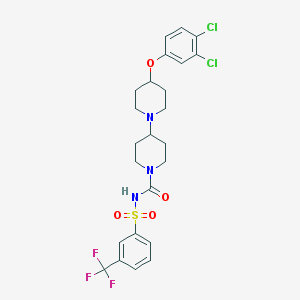 CHEMBL2171045 CHEMBL2171045 | C24H26Cl2F3N3O4S | 580.444 | 8 / 1 | 5.6 | No |
| 12019 | 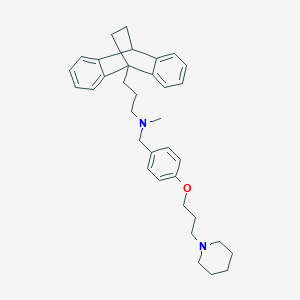 CHEMBL517244 CHEMBL517244 | C35H44N2O | 508.75 | 3 / 0 | 7.8 | No |
| 12039 | 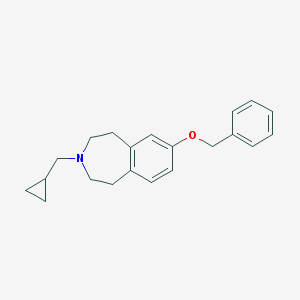 CHEMBL3094114 CHEMBL3094114 | C21H25NO | 307.437 | 2 / 0 | 4.5 | Yes |
| 12133 | 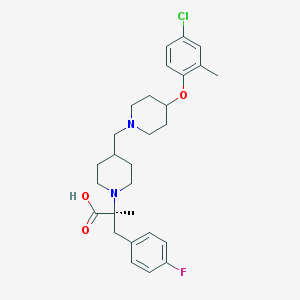 CHEMBL2158791 CHEMBL2158791 | C28H36ClFN2O3 | 503.055 | 6 / 1 | 3.9 | No |
| 548026 | 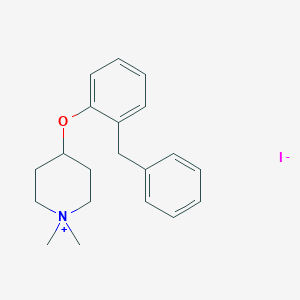 CHEMBL3944629 CHEMBL3944629 | C20H26INO | 423.338 | 2 / 0 | N/A | N/A |
| 13354 | 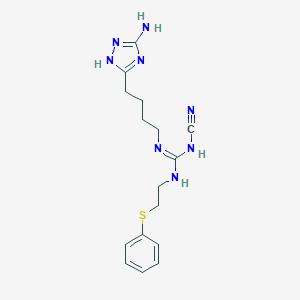 CHEMBL3220649 CHEMBL3220649 | C16H22N8S | 358.468 | 6 / 4 | 2.2 | Yes |
| 14177 | 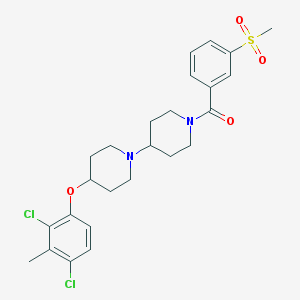 CHEMBL2171021 CHEMBL2171021 | C25H30Cl2N2O4S | 525.485 | 5 / 0 | 5.1 | No |
| 14650 | 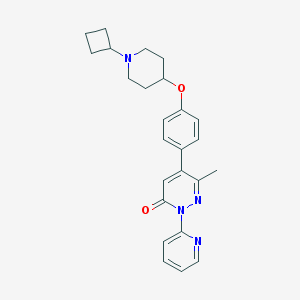 CHEMBL1945848 CHEMBL1945848 | C25H28N4O2 | 416.525 | 5 / 0 | 3.7 | Yes |
| 548049 | 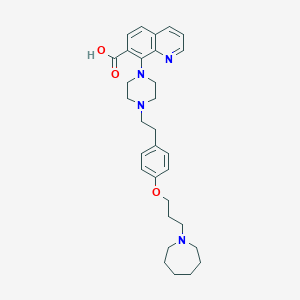 CHEMBL3925977 CHEMBL3925977 | C31H40N4O3 | 516.686 | 7 / 1 | 2.9 | No |
| 14979 | 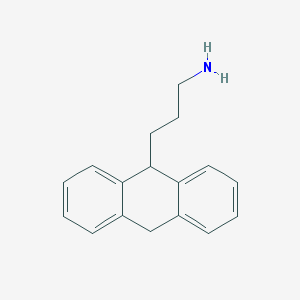 CHEMBL158780 CHEMBL158780 | C17H19N | 237.346 | 1 / 1 | 3.4 | Yes |
| 15143 | 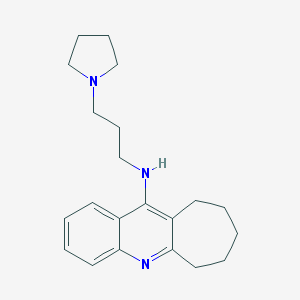 CHEMBL66388 CHEMBL66388 | C21H29N3 | 323.484 | 3 / 1 | 4.8 | Yes |
| 15265 | 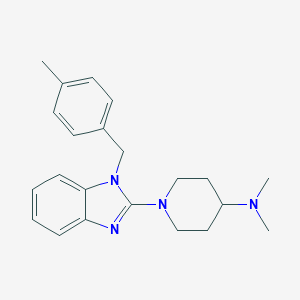 CHEMBL563451 CHEMBL563451 | C22H28N4 | 348.494 | 3 / 0 | 4.4 | Yes |
| 521910 | 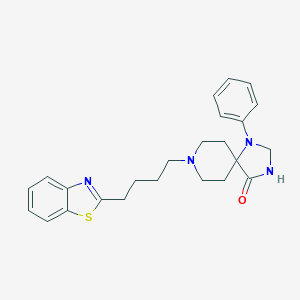 CHEMBL3819567 CHEMBL3819567 | C24H28N4OS | 420.575 | 5 / 1 | 4.8 | Yes |
| 15466 | 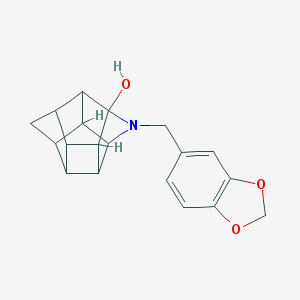 CHEMBL2205829 CHEMBL2205829 | C19H19NO3 | 309.365 | 4 / 1 | 2.2 | Yes |
| 15689 | 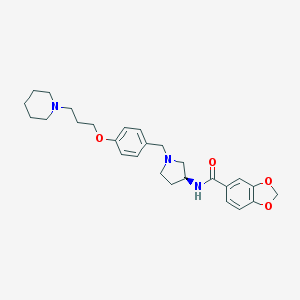 CHEMBL3094128 CHEMBL3094128 | C27H35N3O4 | 465.594 | 6 / 1 | 4.0 | Yes |
| 15779 | 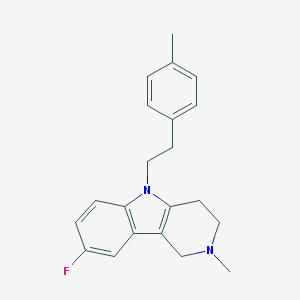 CHEMBL603049 CHEMBL603049 | C21H23FN2 | 322.427 | 2 / 0 | 4.3 | Yes |
| 16077 | 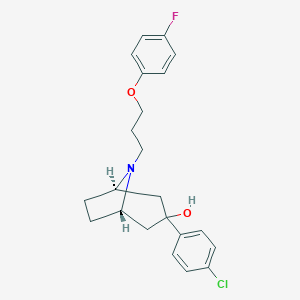 CHEMBL1946122 CHEMBL1946122 | C22H25ClFNO2 | 389.895 | 4 / 1 | 4.8 | Yes |
| 442286 | 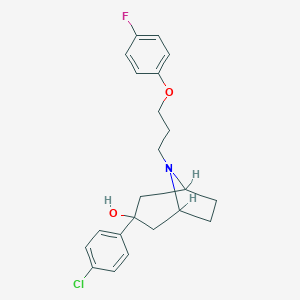 CHEMBL3321789 CHEMBL3321789 | C22H25ClFNO2 | 389.895 | 4 / 1 | 4.8 | Yes |
| 521932 | 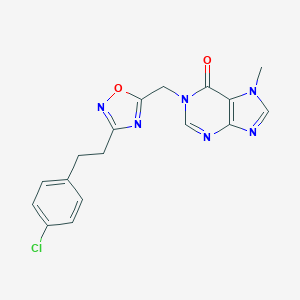 AM-0902 AM-0902 | C17H15ClN6O2 | 370.797 | 6 / 0 | 2.3 | Yes |
| 16117 |  CHEMBL1767165 CHEMBL1767165 | C40H52N4O3 | 636.881 | 6 / 0 | 7.6 | No |
| 16284 |  CHEMBL64479 CHEMBL64479 | C23H36N2O3 | 388.552 | 4 / 0 | 4.0 | Yes |
| 16462 |  CHEMBL540982 CHEMBL540982 | C19H20FNS | 313.434 | 3 / 0 | 5.3 | No |
| 16637 |  CHEMBL60559 CHEMBL60559 | C24H31N3O3S | 441.59 | 6 / 1 | 2.5 | Yes |
| 16723 | 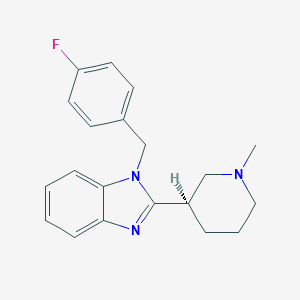 CHEMBL1095819 CHEMBL1095819 | C20H22FN3 | 323.415 | 3 / 0 | 3.7 | Yes |
| 16725 | 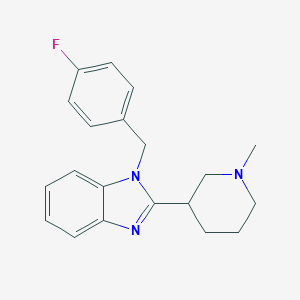 CHEMBL1087089 CHEMBL1087089 | C20H22FN3 | 323.415 | 3 / 0 | 3.7 | Yes |
| 16780 | 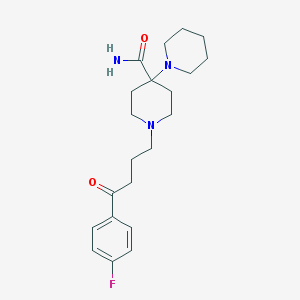 Pipamperone Pipamperone | C21H30FN3O2 | 375.488 | 5 / 1 | 2.0 | Yes |
| 16876 | 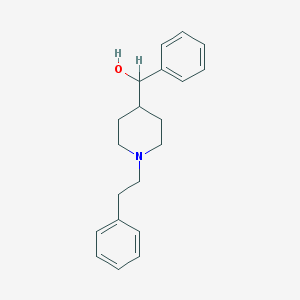 Glemanserin Glemanserin | C20H25NO | 295.426 | 2 / 1 | 3.8 | Yes |
| 548081 | 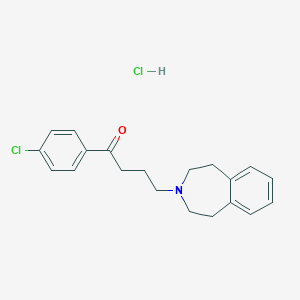 CHEMBL3952251 CHEMBL3952251 | C20H23Cl2NO | 364.31 | 2 / 1 | N/A | N/A |
| 17144 | 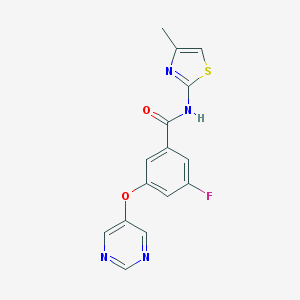 CHEMBL2440659 CHEMBL2440659 | C15H11FN4O2S | 330.337 | 7 / 1 | 2.4 | Yes |
| 521986 | 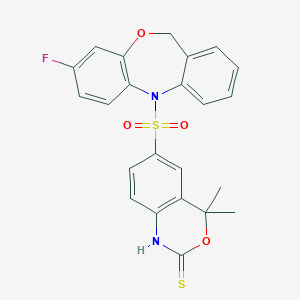 CHEMBL3775473 CHEMBL3775473 | C23H19FN2O4S2 | 470.533 | 7 / 1 | 4.2 | Yes |
| 17386 | 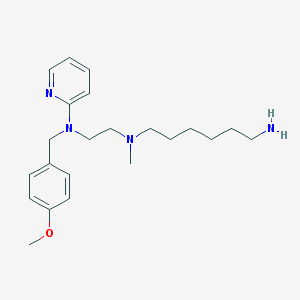 CHEMBL21278 CHEMBL21278 | C22H34N4O | 370.541 | 5 / 1 | 3.8 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417