

You can:
| Name | C-X-C chemokine receptor type 3 |
|---|---|
| Species | Homo sapiens (Human) |
| Gene | CXCR3 |
| Synonym | MigR IP-10 receptor Interferon-inducible protein 10 receptor GPR9 G protein-coupled receptor 9 [ Show all ] |
| Disease | Inflammatory disease Autoimmune diabetes Inflammatory disorder Rheumatoid arthritis Psoriatic disorder |
| Length | 368 |
| Amino acid sequence | MVLEVSDHQVLNDAEVAALLENFSSSYDYGENESDSCCTSPPCPQDFSLNFDRAFLPALYSLLFLLGLLGNGAVAAVLLSRRTALSSTDTFLLHLAVADTLLVLTLPLWAVDAAVQWVFGSGLCKVAGALFNINFYAGALLLACISFDRYLNIVHATQLYRRGPPARVTLTCLAVWGLCLLFALPDFIFLSAHHDERLNATHCQYNFPQVGRTALRVLQLVAGFLLPLLVMAYCYAHILAVLLVSRGQRRLRAMRLVVVVVVAFALCWTPYHLVVLVDILMDLGALARNCGRESRVDVAKSVTSGLGYMHCCLNPLLYAFVGVKFRERMWMLLLRLGCPNQRGLQRQPSSSRRDSSWSETSEASYSGL |
| UniProt | P49682 |
| Protein Data Bank | N/A |
| GPCR-HGmod model | P49682 |
| 3D structure model | This predicted structure model is from GPCR-EXP P49682. |
| BioLiP | N/A |
| Therapeutic Target Database | T25315 |
| ChEMBL | CHEMBL4441 |
| IUPHAR | 70 |
| DrugBank | N/A |
You can:
| GLASS ID | Molecule | Formula | Molecular weight | H-bond acceptor / donor | XlogP | Lipinski's druglikeness |
|---|---|---|---|---|---|---|
| 29 | 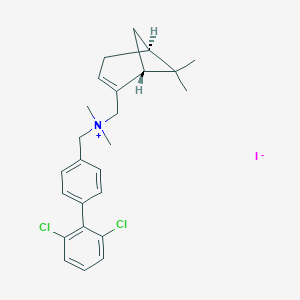 CHEMBL2181460 CHEMBL2181460 | C25H30Cl2IN | 542.326 | 1 / 0 | N/A | No |
| 31 | 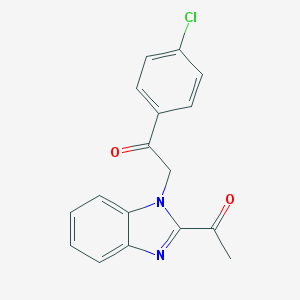 NSC674257 NSC674257 | C17H13ClN2O2 | 312.753 | 3 / 0 | 3.6 | Yes |
| 650 | 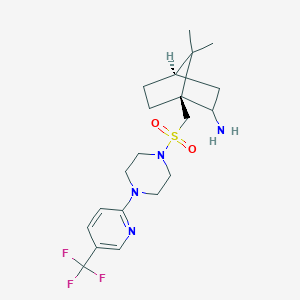 CHEMBL482369 CHEMBL482369 | C20H29F3N4O2S | 446.533 | 9 / 1 | 2.7 | Yes |
| 1084 | 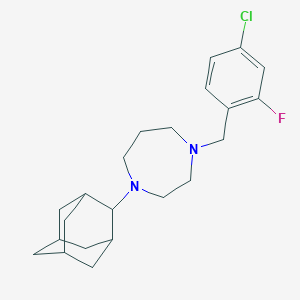 CHEMBL472484 CHEMBL472484 | C22H30ClFN2 | 376.944 | 3 / 0 | 5.0 | Yes |
| 1880 |  CHEMBL200759 CHEMBL200759 | C30H33ClFN5O3 | 566.074 | 5 / 3 | 4.7 | No |
| 2373 |  CHEMBL2205078 CHEMBL2205078 | C26H33N2+ | 373.564 | 0 / 0 | 5.2 | No |
| 3289 |  CHEMBL523249 CHEMBL523249 | C20H32N2O | 316.489 | 3 / 1 | 4.0 | Yes |
| 3482 |  CHEMBL464921 CHEMBL464921 | C21H27N3O3S2 | 433.585 | 7 / 0 | 3.3 | Yes |
| 4296 |  CHEMBL480601 CHEMBL480601 | C21H28F3N3O3S | 459.528 | 9 / 1 | 3.3 | Yes |
| 4788 |  CHEMBL372529 CHEMBL372529 | C30H31Cl2F2N5O3 | 618.507 | 6 / 3 | 5.4 | No |
| 5533 |  CHEMBL1681868 CHEMBL1681868 | C25H33Cl2N5O | 490.473 | 5 / 1 | 4.5 | Yes |
| 5693 |  CHEMBL1085258 CHEMBL1085258 | C29H28N2O4S | 500.613 | 5 / 1 | 4.8 | No |
| 6003 |  CHEMBL372528 CHEMBL372528 | C29H31Cl2N5O3 | 568.499 | 4 / 3 | 4.7 | No |
| 6065 |  CHEMBL2205715 CHEMBL2205715 | C18H24IN | 381.301 | 1 / 1 | 5.3 | No |
| 6118 |  CHEMBL398003 CHEMBL398003 | C23H31N3O3 | 397.519 | 4 / 3 | 0.8 | Yes |
| 6176 |  CHEMBL256585 CHEMBL256585 | C32H34F3N3O5S | 629.695 | 9 / 0 | 5.9 | No |
| 6519 | 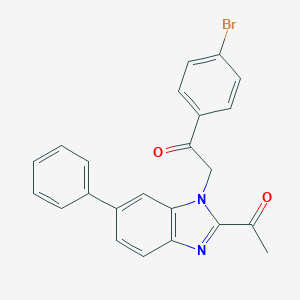 CHEMBL409900 CHEMBL409900 | C23H17BrN2O2 | 433.305 | 3 / 0 | 5.3 | No |
| 6537 | 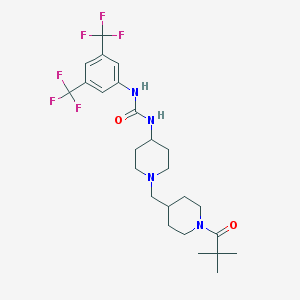 CHEMBL252503 CHEMBL252503 | C25H34F6N4O2 | 536.563 | 9 / 2 | 4.9 | No |
| 6979 | 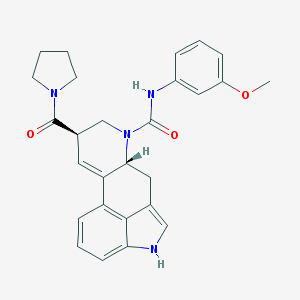 CHEMBL1809019 CHEMBL1809019 | C27H28N4O3 | 456.546 | 3 / 2 | 2.8 | Yes |
| 536152 | 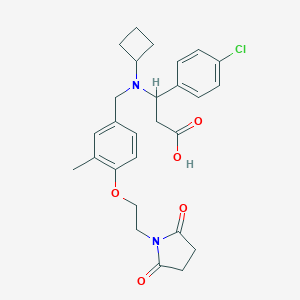 CHEMBL3898394 CHEMBL3898394 | C27H31ClN2O5 | 499.004 | 6 / 1 | 1.3 | Yes |
| 7393 |  CHEMBL395314 CHEMBL395314 | C23H33N3O2 | 383.536 | 3 / 2 | 3.9 | Yes |
| 8109 |  CHEMBL512663 CHEMBL512663 | C23H32ClFN2 | 390.971 | 3 / 1 | 5.5 | No |
| 8572 |  CHEMBL2181454 CHEMBL2181454 | C19H27ClN+ | 304.882 | 0 / 0 | 4.5 | Yes |
| 8620 |  CHEMBL250657 CHEMBL250657 | C24H29Cl2N3S | 462.477 | 4 / 1 | 6.8 | No |
| 10854 | 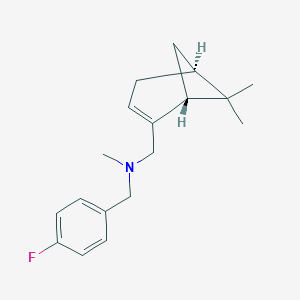 CHEMBL2205710 CHEMBL2205710 | C18H24FN | 273.395 | 2 / 0 | 3.9 | Yes |
| 11145 | 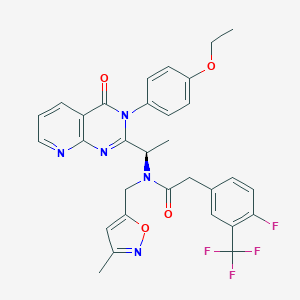 CHEMBL1077823 CHEMBL1077823 | C31H27F4N5O4 | 609.582 | 11 / 0 | 4.9 | No |
| 464275 | 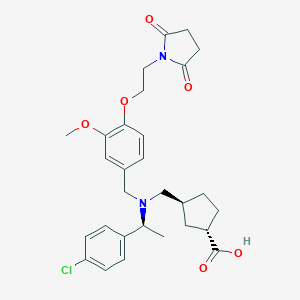 CHEMBL3697136 CHEMBL3697136 | C29H35ClN2O6 | 543.057 | 7 / 1 | 1.5 | No |
| 464276 | 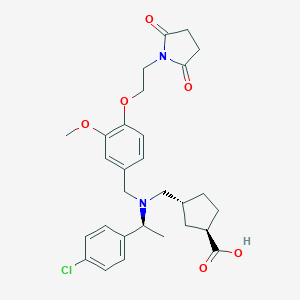 CHEMBL3697135 CHEMBL3697135 | C29H35ClN2O6 | 543.057 | 7 / 1 | 1.5 | No |
| 464277 |  CHEMBL3697138 CHEMBL3697138 | C29H35ClN2O6 | 543.057 | 7 / 1 | 1.5 | No |
| 464278 |  CHEMBL3697137 CHEMBL3697137 | C29H35ClN2O6 | 543.057 | 7 / 1 | 1.5 | No |
| 12025 |  CHEMBL1921875 CHEMBL1921875 | C24H33Cl2N9O3 | 566.488 | 10 / 4 | 2.9 | No |
| 12747 |  CHEMBL395748 CHEMBL395748 | C32H25F3N4O4 | 586.571 | 9 / 1 | 4.8 | No |
| 12798 |  CHEMBL196248 CHEMBL196248 | C30H42N4O2 | 490.692 | 4 / 0 | 6.5 | No |
| 12860 |  CHEMBL1086236 CHEMBL1086236 | C24H24ClN3O3S | 469.984 | 5 / 1 | 3.7 | Yes |
| 13152 |  CHEMBL2181448 CHEMBL2181448 | C25H31IN+ | 472.434 | 0 / 0 | 6.1 | No |
| 557652 |  BDBM228192 BDBM228192 | C23H26F3N7O2S | 521.563 | 11 / 1 | 1.7 | No |
| 13215 |  CHEMBL1681858 CHEMBL1681858 | C24H29Cl2N5O2 | 490.429 | 5 / 1 | 3.2 | Yes |
| 16065 |  CHEMBL272287 CHEMBL272287 | C17H16BrN3O2 | 374.238 | 3 / 1 | 3.1 | Yes |
| 16914 |  CHEMBL1083316 CHEMBL1083316 | C26H23ClN2O3S2 | 511.051 | 5 / 1 | 5.2 | No |
| 16999 |  CHEMBL1809039 CHEMBL1809039 | C30H35N5O2 | 497.643 | 3 / 1 | 2.8 | Yes |
| 17518 |  CHEMBL2205080 CHEMBL2205080 | C25H30N2 | 358.529 | 1 / 0 | 5.2 | No |
| 18405 |  CHEMBL517024 CHEMBL517024 | C23H29N3O3S | 427.563 | 6 / 0 | 3.0 | Yes |
| 18506 |  CHEMBL1085509 CHEMBL1085509 | C22H16F2N6O2S | 466.467 | 9 / 1 | 3.2 | Yes |
| 18988 |  CHEMBL271423 CHEMBL271423 | C29H32F4N4O3S | 592.654 | 9 / 0 | 5.2 | No |
| 19547 |  CHEMBL519434 CHEMBL519434 | C21H28F3N3O3S | 459.528 | 9 / 1 | 3.3 | Yes |
| 548118 |  CHEMBL3900040 CHEMBL3900040 | C36H40N2O7 | 612.723 | 8 / 1 | 2.7 | No |
| 20068 |  CHEMBL395771 CHEMBL395771 | C33H31FN4O2 | 534.635 | 5 / 0 | 5.8 | No |
| 548119 |  CHEMBL3938584 CHEMBL3938584 | C26H32N2O5 | 452.551 | 6 / 1 | 2.9 | Yes |
| 20993 |  CHEMBL1809024 CHEMBL1809024 | C24H24N4O2S | 432.542 | 3 / 2 | 2.5 | Yes |
| 21034 |  CHEMBL2181442 CHEMBL2181442 | C25H31BrN+ | 425.434 | 0 / 0 | 6.1 | No |
| 21202 |  CHEMBL3116473 CHEMBL3116473 | C25H29Cl2FN6O | 519.446 | 8 / 0 | 5.4 | No |
| 548127 |  CHEMBL3981260 CHEMBL3981260 | C32H34N6O5 | 582.661 | 9 / 1 | 3.2 | No |
| 21481 |  CHEMBL1921861 CHEMBL1921861 | C24H33Cl2N7O2 | 522.475 | 8 / 3 | 3.4 | No |
| 21782 |  CID 44443801 CID 44443801 | C26H29F6N3O2 | 529.527 | 9 / 0 | 5.3 | No |
| 22280 |  CHEMBL578189 CHEMBL578189 | C30H29F4N5O3S | 615.648 | 10 / 0 | 5.0 | No |
| 22810 |  CHEMBL253247 CHEMBL253247 | C26H38N4O3 | 454.615 | 4 / 2 | 2.8 | Yes |
| 23152 |  CHEMBL2205712 CHEMBL2205712 | C18H24BrN | 334.301 | 1 / 0 | 4.5 | Yes |
| 23383 |  CHEMBL470431 CHEMBL470431 | C24H34ClFN2 | 404.998 | 3 / 1 | 5.8 | No |
| 23765 |  CHEMBL1809017 CHEMBL1809017 | C26H25FN4O2 | 444.51 | 3 / 2 | 2.9 | Yes |
| 23782 |  CHEMBL1077817 CHEMBL1077817 | C34H28F6N4O3 | 654.613 | 11 / 0 | 6.5 | No |
| 24415 |  CHEMBL436826 CHEMBL436826 | C33H29F3N4O3 | 586.615 | 8 / 0 | 5.6 | No |
| 24515 |  CHEMBL497396 CHEMBL497396 | C26H31BrN2O3 | 499.449 | 4 / 0 | 3.8 | Yes |
| 24611 |  CHEMBL403491 CHEMBL403491 | C28H30F4N4O3S | 578.627 | 9 / 0 | 4.6 | No |
| 24627 |  CHEMBL272742 CHEMBL272742 | C16H16BrN3 | 330.229 | 1 / 1 | 3.7 | Yes |
| 25015 |  CHEMBL459951 CHEMBL459951 | C21H30F3N3O3S | 461.544 | 9 / 1 | 3.2 | Yes |
| 25036 |  CHEMBL495531 CHEMBL495531 | C23H23BrF2N2O2 | 477.35 | 5 / 1 | 4.0 | Yes |
| 25123 |  CHEMBL2205056 CHEMBL2205056 | C25H30Cl2IN | 542.326 | 1 / 0 | N/A | No |
| 25189 |  CHEMBL269811 CHEMBL269811 | C18H15BrN2O3 | 387.233 | 4 / 0 | 3.6 | Yes |
| 25569 |  CHEMBL1921877 CHEMBL1921877 | C26H36Cl2N8O2 | 563.528 | 8 / 2 | 4.4 | No |
| 25708 |  CHEMBL380594 CHEMBL380594 | C28H31Cl2N5O3S | 588.548 | 5 / 3 | 4.9 | No |
| 26384 |  CHEMBL2205077 CHEMBL2205077 | C25H32INO | 489.441 | 2 / 0 | N/A | N/A |
| 522368 |  SCHEMBL18725755 SCHEMBL18725755 | C42H45F3N4O6 | 758.839 | 9 / 3 | N/A | No |
| 522369 |  SCHEMBL18725707 SCHEMBL18725707 | C42H45F3N4O6 | 758.839 | 9 / 3 | N/A | No |
| 26743 | 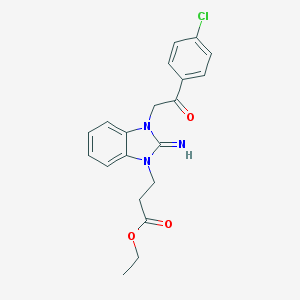 CHEMBL271081 CHEMBL271081 | C20H20ClN3O3 | 385.848 | 4 / 1 | 3.2 | Yes |
| 27447 | 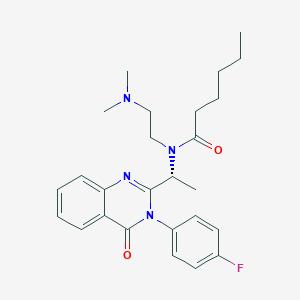 CHEMBL231589 CHEMBL231589 | C26H33FN4O2 | 452.574 | 5 / 0 | 4.4 | Yes |
| 548191 | 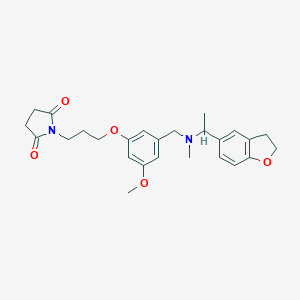 CHEMBL3904870 CHEMBL3904870 | C26H32N2O5 | 452.551 | 6 / 0 | 2.9 | Yes |
| 27864 | 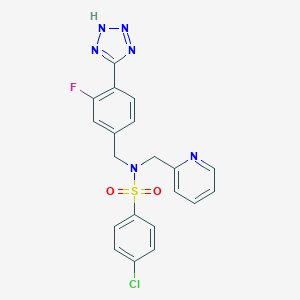 CHEMBL1082623 CHEMBL1082623 | C20H16ClFN6O2S | 458.896 | 8 / 1 | 3.0 | Yes |
| 28157 | 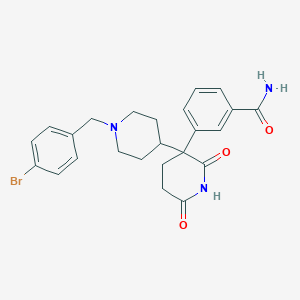 CHEMBL456904 CHEMBL456904 | C24H26BrN3O3 | 484.394 | 4 / 2 | 2.7 | Yes |
| 28245 | 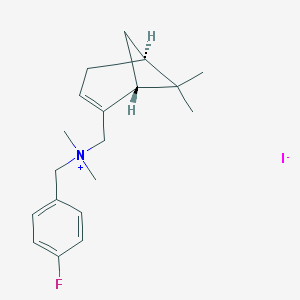 CHEMBL2205694 CHEMBL2205694 | C19H27FIN | 415.335 | 2 / 0 | N/A | N/A |
| 28405 | 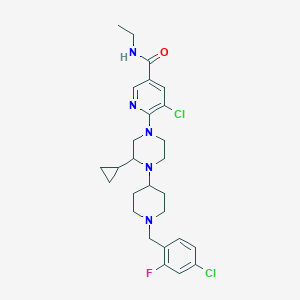 CHEMBL1681884 CHEMBL1681884 | C27H34Cl2FN5O | 534.501 | 6 / 1 | 5.0 | No |
| 30269 | 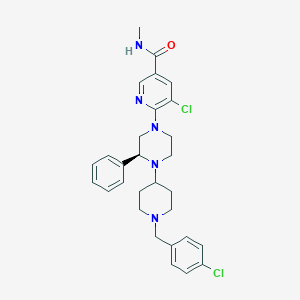 CHEMBL1681876 CHEMBL1681876 | C29H33Cl2N5O | 538.517 | 5 / 1 | 5.3 | No |
| 522465 |  SCHEMBL18725749 SCHEMBL18725749 | C43H47F3N4O6 | 772.866 | 9 / 3 | N/A | No |
| 30499 |  CHEMBL1921873 CHEMBL1921873 | C26H38Cl2N8O3S | 613.603 | 10 / 3 | 3.6 | No |
| 536772 |  CHEMBL3914758 CHEMBL3914758 | C27H32ClN3O5 | 514.019 | 6 / 1 | 1.5 | No |
| 536784 |  CHEMBL3889627 CHEMBL3889627 | C23H27N5O3S | 453.561 | 5 / 2 | 2.6 | Yes |
| 31112 | 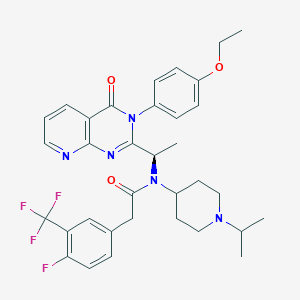 CHEMBL1077828 CHEMBL1077828 | C34H37F4N5O3 | 639.696 | 10 / 0 | 5.9 | No |
| 31445 | 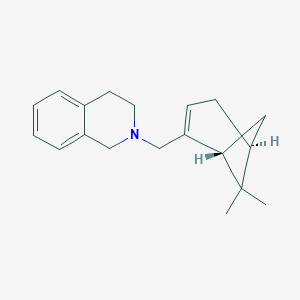 CHEMBL2205709 CHEMBL2205709 | C19H25N | 267.416 | 1 / 0 | 3.8 | Yes |
| 31786 | 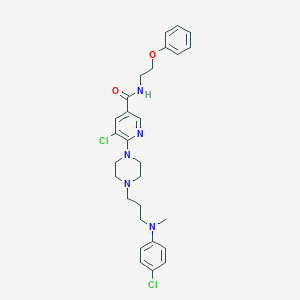 CHEMBL569336 CHEMBL569336 | C28H33Cl2N5O2 | 542.505 | 6 / 1 | 5.5 | No |
| 548249 | 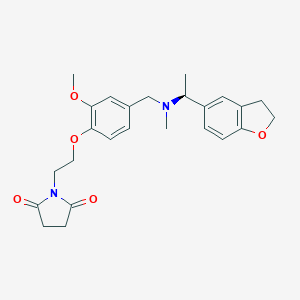 CHEMBL3892445 CHEMBL3892445 | C25H30N2O5 | 438.524 | 6 / 0 | 2.5 | Yes |
| 548250 |  CHEMBL3947168 CHEMBL3947168 | C25H30N2O5 | 438.524 | 6 / 0 | 2.5 | Yes |
| 548251 |  CHEMBL3916901 CHEMBL3916901 | C25H30N2O5 | 438.524 | 6 / 0 | 2.5 | Yes |
| 32964 |  CHEMBL522551 CHEMBL522551 | C23H24BrFN2O2 | 459.359 | 4 / 1 | 3.9 | Yes |
| 34317 |  CHEMBL253733 CHEMBL253733 | C27H36F4N4O2 | 524.605 | 7 / 2 | 4.4 | No |
| 34469 | 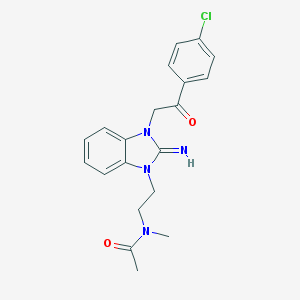 CHEMBL270222 CHEMBL270222 | C20H21ClN4O2 | 384.864 | 3 / 1 | 2.6 | Yes |
| 34625 | 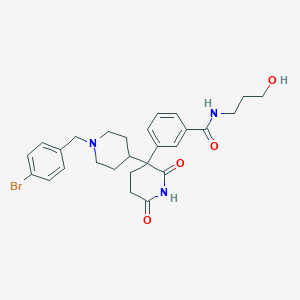 CHEMBL514259 CHEMBL514259 | C27H32BrN3O4 | 542.474 | 5 / 3 | 2.8 | No |
| 35570 | 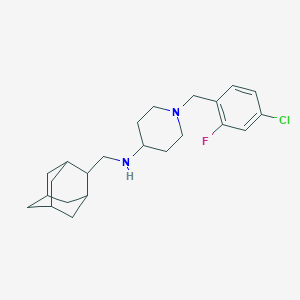 CHEMBL470839 CHEMBL470839 | C23H32ClFN2 | 390.971 | 3 / 1 | 5.9 | No |
| 35721 | 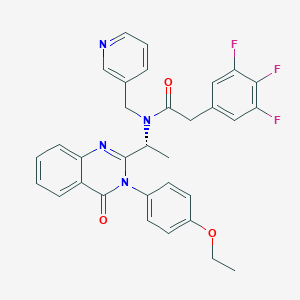 CHEMBL1077816 CHEMBL1077816 | C32H27F3N4O3 | 572.588 | 8 / 0 | 5.0 | No |
| 548304 | 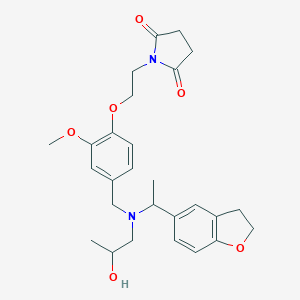 CHEMBL3922202 CHEMBL3922202 | C27H34N2O6 | 482.577 | 7 / 1 | 2.3 | Yes |
| 36134 | 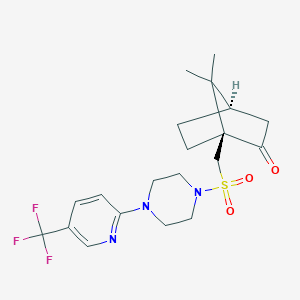 CHEMBL464081 CHEMBL464081 | C20H26F3N3O3S | 445.501 | 9 / 0 | 2.7 | Yes |
| 36136 | 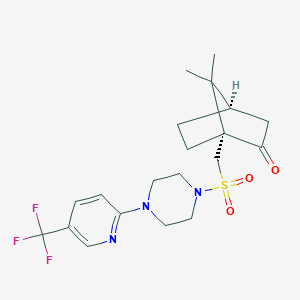 CHEMBL459742 CHEMBL459742 | C20H26F3N3O3S | 445.501 | 9 / 0 | 2.7 | Yes |
zhanglab![]() zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417
zhanggroup.org | +65-6601-1241 | Computing 1, 13 Computing Drive, Singapore 117417